Difference between revisions of "Part:BBa K3349007"
Kristiturton (Talk | contribs) |
Kristiturton (Talk | contribs) |
||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3349007 short</partinfo> | <partinfo>BBa_K3349007 short</partinfo> | ||
| − | This Pectin Lyase is a hypothetical protein from <i> Paenibaciullus amylolyticus </i>. With Pnl and PelC, PelB is able to cleave and therefore breakdown pectin or homogalacturonan substrates. This protein has been characterized to cleave methylated pectin (at unmethylated sites) and polygalacturonic acid [2]. A hexahistidine tag was added onto the C-terminus for nickel affinity purification. For more information, please visit our website https://2020.igem.org/Team:Lethbridge_HS. | + | This Pectin Lyase is a hypothetical protein from <i> Paenibaciullus amylolyticus </i>. With Pnl and PelC, PelB is able to cleave and therefore breakdown pectin or homogalacturonan substrates. This protein has been characterized to cleave methylated pectin (at unmethylated sites) and polygalacturonic acid [2]. Studies by Keggi and Doran-Peterson, PelB has a Km of 50.1+/- 140 mg/ml and a Vmax of 745 +/-2024 IU/mg with a 80% methylated substrate (0 and 55% methylation was also studied). The optimal temperature for PelB activity was determined to be 70°C [1]. |
| + | |||
| + | A hexahistidine tag was added onto the C-terminus for nickel affinity purification. For more information, please visit our website https://2020.igem.org/Team:Lethbridge_HS. | ||
<html> | <html> | ||
Revision as of 23:59, 30 September 2020
Pectin Lyase PelB
This Pectin Lyase is a hypothetical protein from Paenibaciullus amylolyticus . With Pnl and PelC, PelB is able to cleave and therefore breakdown pectin or homogalacturonan substrates. This protein has been characterized to cleave methylated pectin (at unmethylated sites) and polygalacturonic acid [2]. Studies by Keggi and Doran-Peterson, PelB has a Km of 50.1+/- 140 mg/ml and a Vmax of 745 +/-2024 IU/mg with a 80% methylated substrate (0 and 55% methylation was also studied). The optimal temperature for PelB activity was determined to be 70°C [1].
A hexahistidine tag was added onto the C-terminus for nickel affinity purification. For more information, please visit our website https://2020.igem.org/Team:Lethbridge_HS.
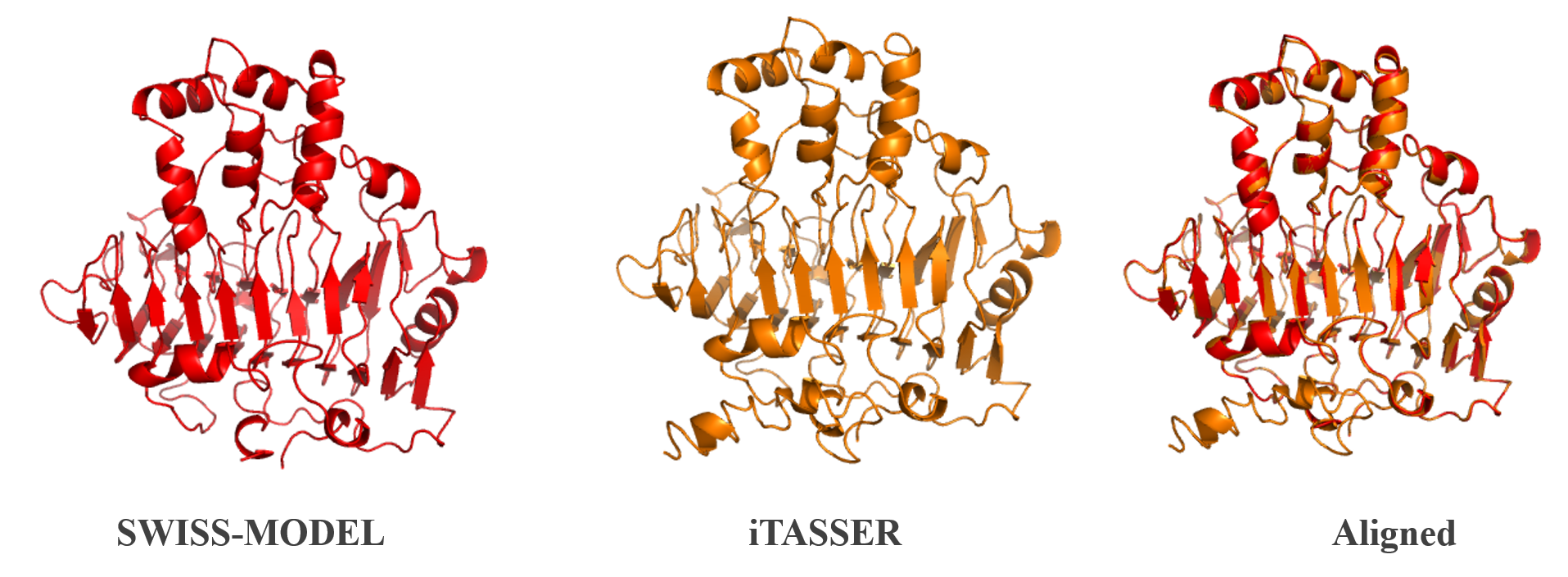
Figure 2:Homology modelling of PelB from P. amylolyticus . The protein structure was done using iTASSER [2] and SWISS-Model [3]. The two structures were then aligned and compared. A more in depth analysis can be found on our webpage.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 333
Illegal BamHI site found at 1218 - 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
