Difference between revisions of "Part:BBa K1150001"
| (13 intermediate revisions by 4 users not shown) | |||
| Line 25: | Line 25: | ||
Virus Protein 16 (VP16) is a transcription factor encoded by the UL48 gene of Herpes simplex virus-1 (HSV-1). | Virus Protein 16 (VP16) is a transcription factor encoded by the UL48 gene of Herpes simplex virus-1 (HSV-1). | ||
| − | Through complex formation with cellular host factors VP16 can bind to a common regulatory element in the upstream | + | Through complex formation with cellular host factors VP16 can bind to a common regulatory element in the upstream promoter region of immediate-early genes [1]. Due to the transactivating function of VP-16 these genes can then be expressed. |
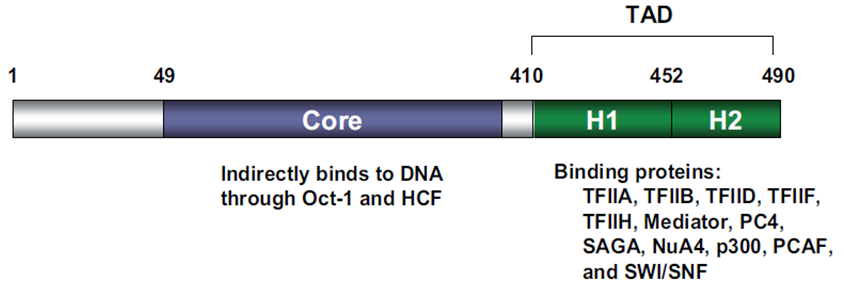
| − | VP16 consists of 490 amino acids with two very important functional domains: a core domain in its central region which is necessary for the indirect DNA binding and a carboxy-terminal transactivation domain | + | VP16 consists of 490 amino acids with two very important functional domains: a core domain in its central region which is necessary for the indirect DNA binding and a carboxy-terminal transactivation domain [2], [3]; see Fig.1. The transactivation domain (TAD) of VP16 can be fused to a DNA-binding domain (DBD) of another protein in order to gain expression of a desired target gene [1]. |
| − | [[File:Picture1VP16_Freigem_2013.png | + | [[File:Picture1VP16_Freigem_2013.png|700px|'''Fig. 1:''' Domain structure of VP16. The different functional domains of VP16 are shown. Taken from Hirai ''et al.'', 2010.]] |
| + | |||
| + | '''Fig. 1:''' Domain structure of VP16. The different functional domains of VP16 are shown. Taken from Hirai ''et al.'', 2010. | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| Line 35: | Line 37: | ||
==Transactivation domain (TAD) of VP16== | ==Transactivation domain (TAD) of VP16== | ||
| − | The TAD of VP16 is one of the most efficient TADs. It is widely fused to host transcription factors in order to increase their activity or, next to this, to other transcription factors to study the mechanism of gene regulation [ | + | The TAD of VP16 is one of the most efficient TADs. It is widely fused to host transcription factors in order to increase their activity or, next to this, to other transcription factors to study the mechanism of gene regulation [1]. |
| − | In our case the TAD of VP16 was cloned behind a double mutated Cas9 gene (which encodes for a non-cutting | + | In our case the TAD of VP16 was cloned behind a double mutated Cas9 gene (which encodes for a non-cutting dCas9 protein with specific DNA binding properties) to activate our genes of interest. For more information see BBa_K1150019 and BBa_K1150020. |
==Biology== | ==Biology== | ||
VP16 is originally a part of the virus particle from HSV and is released into an animal cell while infection. VP16 first binds via its core domain to the host nuclear protein which then recruits another host nuclear protein, Oct-1, to form a functional protein complex. This complex then binds to | VP16 is originally a part of the virus particle from HSV and is released into an animal cell while infection. VP16 first binds via its core domain to the host nuclear protein which then recruits another host nuclear protein, Oct-1, to form a functional protein complex. This complex then binds to | ||
| − | its target DNA sequence in the | + | its target DNA sequence in the promoter region of immediate-early genes where VP16 activates these |
genes through interactions between its TAD and many different | genes through interactions between its TAD and many different | ||
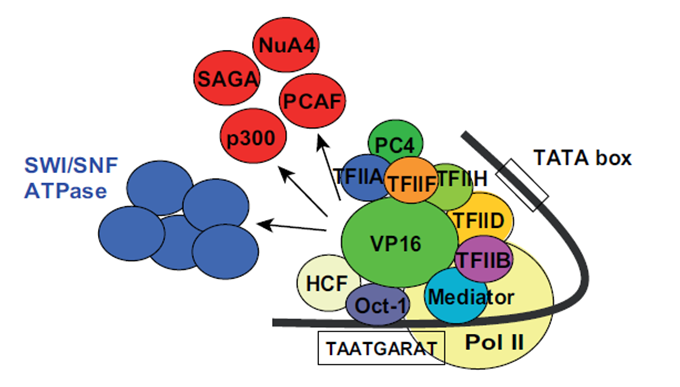
| − | transcription factors and other proteins, such as histone acetyltransferases; see Fig.2. Through these multiple interactions the transactivation domain of VP16 facilitates the assembly of the pre-initiation complex [ | + | transcription factors and other proteins, such as histone acetyltransferases; see Fig.2. Through these multiple interactions the transactivation domain of VP16 facilitates the assembly of the pre-initiation complex [5], [1]. Also other TAD containing proteins share this magnitude of binding partners but VP16 is one of the most effective once [1]. |
| + | |||
| + | [[File:Picture2VP16_Freigem2013.png]] | ||
'''Fig. 2:''' Interaction partners of VP16. Proteins are shown that interact with VP16 in the phase of gene activation. Taken from Hirai ''et al.'', 2010 | '''Fig. 2:''' Interaction partners of VP16. Proteins are shown that interact with VP16 in the phase of gene activation. Taken from Hirai ''et al.'', 2010 | ||
| − | == | + | <span class='h3bb'> |
| + | ==Sequence and Features== | ||
| + | </span> | ||
| + | <partinfo>BBa_K1150001 SequenceAndFeatures</partinfo> | ||
| + | <br> | ||
| + | <html><!--- Please copy this table containing parameters for BBa_ at the end of the parametrs section ahead of the references. ---><style type="text/css">table#AutoAnnotator {border:1px solid black; width:100%; border-collapse:collapse;} th#AutoAnnotatorHeader { border:1px solid black; width:100%; background-color: rgb(221, 221, 221);} td.AutoAnnotator1col { width:100%; border:1px solid black; } span.AutoAnnotatorSequence { font-family:'Courier New', Arial; } td.AutoAnnotatorSeqNum { text-align:right; width:2%; } td.AutoAnnotatorSeqSeq { width:98% } td.AutoAnnotatorSeqFeat1 { width:3% } td.AutoAnnotatorSeqFeat2a { width:27% } td.AutoAnnotatorSeqFeat2b { width:97% } td.AutoAnnotatorSeqFeat3 { width:70% } table.AutoAnnotatorNoBorder { border:0px; width:100%; border-collapse:collapse; } table.AutoAnnotatorWithBorder { border:1px solid black; width:100%; border-collapse:collapse; } td.AutoAnnotatorOuterAmino { border:0px solid black; width:20% } td.AutoAnnotatorInnerAmino { border:1px solid black; width:50% } td.AutoAnnotatorAminoCountingOuter { border:1px solid black; width:40%; } td.AutoAnnotatorBiochemParOuter { border:1px solid black; width:60%; } td.AutoAnnotatorAminoCountingInner1 { width: 7.5% } td.AutoAnnotatorAminoCountingInner2 { width:62.5% } td.AutoAnnotatorAminoCountingInner3 { width:30% } td.AutoAnnotatorBiochemParInner1 { width: 5% } td.AutoAnnotatorBiochemParInner2 { width:55% } td.AutoAnnotatorBiochemParInner3 { width:40% } td.AutoAnnotatorCodonUsage1 { width: 3% } td.AutoAnnotatorCodonUsage2 { width:14.2% } td.AutoAnnotatorCodonUsage3 { width:13.8% } td.AutoAnnotatorAlignment1 { width: 3% } td.AutoAnnotatorAlignment2 { width: 10% } td.AutoAnnotatorAlignment3 { width: 87% } td.AutoAnnotatorLocalizationOuter {border:1px solid black; width:40%} td.AutoAnnotatorGOOuter {border:1px solid black; width:60%} td.AutoAnnotatorLocalization1 { width: 7.5% } td.AutoAnnotatorLocalization2 { width: 22.5% } td.AutoAnnotatorLocalization3 { width: 70% } td.AutoAnnotatorGO1 { width: 5% } td.AutoAnnotatorGO2 { width: 35% } td.AutoAnnotatorGO3 { width: 60% } td.AutoAnnotatorPredFeat1 { width:3% } td.AutoAnnotatorPredFeat2a { width:27% } td.AutoAnnotatorPredFeat3 { width:70% } div.AutoAnnotator_trans { position:absolute; background:rgb(11,140,143); background-color:rgba(11,140,143, 0.8); height:5px; top:100px; } div.AutoAnnotator_sec_helix { position:absolute; background:rgb(102,0,102); background-color:rgba(102,0,102, 0.8); height:5px; top:110px; } div.AutoAnnotator_sec_strand { position:absolute; background:rgb(245,170,26); background-color:rgba(245,170,26, 1); height:5px; top:110px; } div.AutoAnnotator_acc_buried { position:absolute; background:rgb(89,168,15); background-color:rgba(89,168,15, 0.8); height:5px; top:120px; } div.AutoAnnotator_acc_exposed { position:absolute; background:rgb(0, 0, 255); background-color:rgba(0, 0, 255, 0.8); height:5px; top:120px; } div.AutoAnnotator_dis { position:absolute; text-align:center; font-family:Arial,Helvetica,sans-serif; background:rgb(255, 200, 0); background-color:rgba(255, 200, 0, 1); height:16px; width:16px; top:80px; border-radius:50%; } </style><div id='AutoAnnotator_container_1381279325962'><table id="AutoAnnotator"><tr><!-- Time stamp in ms since 1/1/1970 1381279325962 --><th id="AutoAnnotatorHeader" colspan="2">Protein data table for BioBrick <a href="https://parts.igem.org/wiki/index.php?title=Part:BBa_K1150001<!------------------------Enter BioBrick number here------------------------>">BBa_K1150001<!------------------------Enter BioBrick number here------------------------></a> automatically created by the <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">BioBrick-AutoAnnotator</a> version 1.0</th></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Nucleotide sequence</strong> in <strong>RFC 25</strong>, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein)<br><span class="AutoAnnotatorSequence"> <u><i>ATGGCCGGC</i>GCGCGTACG ... TACGGTGGG<i>ACCGGT</i></u></span><br> <strong>ORF</strong> from nucleotide position -8 to 378 (excluding stop-codon)</td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid sequence:</strong> (RFC 25 scars in shown in bold, other sequence features underlined; both given below)<br><span class="AutoAnnotatorSequence"><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqNum">1 <br>101 </td><td class="AutoAnnotatorSeqSeq">MAGARTKNNYGSTIEGLLDLPDDDAPEEAGLAAPRLSFLPAGHTRRLSTAPPTDVSLGDELHLDGEDVAMAHADALDDFDLDMLGDGDSPGPGFTPHDSA<br>PYGALDMADFEFEQMFTDALGIDEYGGTG*</td></tr></table></span></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Sequence features:</strong> (with their position in the amino acid sequence, see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software/FeatureList">list of supported features</a>)<table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorSeqFeat1"></td><td class="AutoAnnotatorSeqFeat2b">None of the supported features appeared in the sequence</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Amino acid composition:</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Ala (A)</td><td class="AutoAnnotatorInnerAmino">16 (12.4%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Arg (R)</td><td class="AutoAnnotatorInnerAmino">4 (3.1%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asn (N)</td><td class="AutoAnnotatorInnerAmino">2 (1.6%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Asp (D)</td><td class="AutoAnnotatorInnerAmino">20 (15.5%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Cys (C)</td><td class="AutoAnnotatorInnerAmino">0 (0.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gln (Q)</td><td class="AutoAnnotatorInnerAmino">1 (0.8%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Glu (E)</td><td class="AutoAnnotatorInnerAmino">8 (6.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Gly (G)</td><td class="AutoAnnotatorInnerAmino">16 (12.4%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">His (H)</td><td class="AutoAnnotatorInnerAmino">4 (3.1%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ile (I)</td><td class="AutoAnnotatorInnerAmino">2 (1.6%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Leu (L)</td><td class="AutoAnnotatorInnerAmino">15 (11.6%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Lys (K)</td><td class="AutoAnnotatorInnerAmino">1 (0.8%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Met (M)</td><td class="AutoAnnotatorInnerAmino">5 (3.9%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Phe (F)</td><td class="AutoAnnotatorInnerAmino">6 (4.7%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Pro (P)</td><td class="AutoAnnotatorInnerAmino">10 (7.8%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Ser (S)</td><td class="AutoAnnotatorInnerAmino">6 (4.7%)</td></tr></table></td><td class="AutoAnnotatorOuterAmino"><table class="AutoAnnotatorWithBorder"><tr><td class="AutoAnnotatorInnerAmino">Thr (T)</td><td class="AutoAnnotatorInnerAmino">8 (6.2%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Trp (W)</td><td class="AutoAnnotatorInnerAmino">0 (0.0%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Tyr (Y)</td><td class="AutoAnnotatorInnerAmino">3 (2.3%)</td></tr><tr><td class="AutoAnnotatorInnerAmino">Val (V)</td><td class="AutoAnnotatorInnerAmino">2 (1.6%)</td></tr></table></td></tr></table></td></tr><tr><td class="AutoAnnotatorAminoCountingOuter"><strong>Amino acid counting</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Total number:</td><td class="AutoAnnotatorAminoCountingInner3">129</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Positively charged (Arg+Lys):</td><td class="AutoAnnotatorAminoCountingInner3">5 (3.9%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Negatively charged (Asp+Glu):</td><td class="AutoAnnotatorAminoCountingInner3">28 (21.7%)</td></tr><tr><td class="AutoAnnotatorAminoCountingInner1"></td><td class="AutoAnnotatorAminoCountingInner2">Aromatic (Phe+His+Try+Tyr):</td><td class="AutoAnnotatorAminoCountingInner3">13 (10.1%)</td></tr></table></td><td class="AutoAnnotatorBiochemParOuter"><strong>Biochemical parameters</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Atomic composition:</td><td class="AutoAnnotatorBiochemParInner3">C<sub>585</sub>H<sub>881</sub>N<sub>153</sub>O<sub>206</sub>S<sub>5</sub></td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Molecular mass [Da]:</td><td class="AutoAnnotatorBiochemParInner3">13513.6</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Theoretical pI:</td><td class="AutoAnnotatorBiochemParInner3">3.85</td></tr><tr><td class="AutoAnnotatorBiochemParInner1"></td><td class="AutoAnnotatorBiochemParInner2">Extinction coefficient at 280 nm [M<sup>-1</sup> cm<sup>-1</sup>]:</td><td class="AutoAnnotatorBiochemParInner3">4470 / 4470 (all Cys red/ox)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges</strong> <input type='button' id='hydrophobicity_charge_button' onclick='show_or_hide_plot_1381279325962()' value='Show'><span id="hydrophobicity_charge_explanation"></span><div id="hydrophobicity_charge_container" style='display:none'><div id="hydrophobicity_charge_placeholder0" style="width:100%;height:150px"></div></div></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Codon usage</strong><table class="AutoAnnotatorNoBorder"><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Organism:</td><td class="AutoAnnotatorCodonUsage3"><i>E. coli</i></td><td class="AutoAnnotatorCodonUsage3"><i>B. subtilis</i></td><td class="AutoAnnotatorCodonUsage3"><i>S. cerevisiae</i></td><td class="AutoAnnotatorCodonUsage3"><i>A. thaliana</i></td><td class="AutoAnnotatorCodonUsage3"><i>P. patens</i></td><td class="AutoAnnotatorCodonUsage3">Mammals</td></tr><tr><td class="AutoAnnotatorCodonUsage1"></td><td class="AutoAnnotatorCodonUsage2">Codon quality (<a href="http://en.wikipedia.org/wiki/Codon_Adaptation_Index">CAI</a>):</td><td class="AutoAnnotatorCodonUsage3">good (0.70)</td><td class="AutoAnnotatorCodonUsage3">good (0.67)</td><td class="AutoAnnotatorCodonUsage3">acceptable (0.50)</td><td class="AutoAnnotatorCodonUsage3">acceptable (0.57)</td><td class="AutoAnnotatorCodonUsage3">excellent (0.81)</td><td class="AutoAnnotatorCodonUsage3">good (0.72)</td></tr></table></td></tr><tr><td class="AutoAnnotator1col" colspan="2"><strong>Alignments</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)<br> There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours.</td></tr><tr><th id='AutoAnnotatorHeader' colspan="2"><strong>Predictions</strong> (obtained from <a href='http://predictprotein.org'>PredictProtein.org</a>)</th></tr><tr><td class="AutoAnnotator1col" colspan="2"> There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours.</td><tr><td class="AutoAnnotator1col" colspan="2"> The BioBrick-AutoAnnotator was created by <a href="http://2013.igem.org/Team:TU-Munich">TU-Munich 2013</a> iGEM team. For more information please see the <a href="http://2013.igem.org/Team:TU-Munich/Results/Software">documentation</a>.<br>If you have any questions, comments or suggestions, please leave us a <a href="http://2013.igem.org/Team:TU-Munich/Results/AutoAnnotator">comment</a>.</td></tr></table></div><br><!-- IMPORTANT: DON'T REMOVE THIS LINE, OTHERWISE NOT SUPPORTED FOR IE BEFORE 9 --><!--[if lte IE 8]><script language="javascript" type="text/javascript" src="excanvas.min.js"></script><![endif]--><script type='text/javascript' src='http://code.jquery.com/jquery-1.10.0.min.js'></script><script type='text/javascript' src='http://2013.igem.org/Team:TU-Munich/Flot.js?action=raw&ctype=text/js'></script><script>function show_or_hide_plot_1381279325962(){hydrophobicity_datapoints = [[2.5,0.12],[3.5,-0.40],[4.5,-1.54],[5.5,-2.16],[6.5,-3.22],[7.5,-2.58],[8.5,-2.52],[9.5,-1.90],[10.5,-1.34],[11.5,0.26],[12.5,-0.18],[13.5,-0.18],[14.5,0.74],[15.5,1.64],[16.5,0.04],[17.5,1.50],[18.5,1.26],[19.5,-0.20],[20.5,-1.66],[21.5,-1.66],[22.5,-2.06],[23.5,-2.06],[24.5,-2.06],[25.5,-2.06],[26.5,-1.00],[27.5,-1.44],[28.5,-0.36],[29.5,0.70],[30.5,1.76],[31.5,1.08],[32.5,0.26],[33.5,0.26],[34.5,-0.26],[35.5,-0.06],[36.5,1.02],[37.5,1.60],[38.5,1.20],[39.5,1.28],[40.5,0.08],[41.5,-0.82],[42.5,-1.40],[43.5,-2.66],[44.5,-1.82],[45.5,-1.34],[46.5,-1.34],[47.5,-0.08],[48.5,0.50],[49.5,-0.58],[50.5,-0.56],[51.5,-1.12],[52.5,-0.64],[53.5,-0.48],[54.5,0.60],[55.5,0.66],[56.5,0.66],[57.5,-0.88],[58.5,0.04],[59.5,-1.36],[60.5,-0.52],[61.5,-0.52],[62.5,0.10],[63.5,-1.36],[64.5,-1.42],[65.5,-1.34],[66.5,-0.28],[67.5,0.18],[68.5,1.24],[69.5,1.30],[70.5,0.82],[71.5,-0.24],[72.5,-0.26],[73.5,0.14],[74.5,0.08],[75.5,-0.98],[76.5,0.28],[77.5,-0.78],[78.5,-0.78],[79.5,-0.78],[80.5,0.30],[81.5,0.50],[82.5,1.12],[83.5,-0.34],[84.5,0.28],[85.5,-0.80],[86.5,-1.72],[87.5,-1.96],[88.5,-1.34],[89.5,-1.58],[90.5,-0.96],[91.5,-0.24],[92.5,-0.06],[93.5,-0.30],[94.5,-0.62],[95.5,-1.24],[96.5,-1.96],[97.5,-1.46],[98.5,-1.46],[99.5,-1.08],[100.5,-0.46],[101.5,0.06],[102.5,0.46],[103.5,0.08],[104.5,0.72],[105.5,1.16],[106.5,0.10],[107.5,-0.10],[108.5,-0.10],[109.5,0.08],[110.5,-0.98],[111.5,-0.98],[112.5,-1.16],[113.5,0.10],[114.5,-0.60],[115.5,-0.60],[116.5,0.46],[117.5,0.84],[118.5,0.20],[119.5,1.24],[120.5,1.24],[121.5,0.18],[122.5,-0.84],[123.5,-0.84],[124.5,-1.82],[125.5,-1.26],[126.5,-0.64]];charge_datapoints = [[2.5,0.20],[3.5,0.20],[4.5,0.40],[5.5,0.40],[6.5,0.40],[7.5,0.20],[8.5,0.20],[9.5,0.00],[10.5,0.00],[11.5,0.00],[12.5,-0.20],[13.5,-0.20],[14.5,-0.20],[15.5,-0.20],[16.5,-0.40],[17.5,-0.20],[18.5,-0.20],[19.5,-0.40],[20.5,-0.60],[21.5,-0.60],[22.5,-0.60],[23.5,-0.60],[24.5,-0.60],[25.5,-0.60],[26.5,-0.40],[27.5,-0.40],[28.5,-0.40],[29.5,-0.20],[30.5,-0.00],[31.5,-0.00],[32.5,0.20],[33.5,0.20],[34.5,0.20],[35.5,0.20],[36.5,0.20],[37.5,-0.00],[38.5,-0.00],[39.5,-0.00],[40.5,0.10],[41.5,0.10],[42.5,0.30],[43.5,0.50],[44.5,0.50],[45.5,0.40],[46.5,0.40],[47.5,0.20],[48.5,-0.00],[49.5,-0.00],[50.5,-0.00],[51.5,-0.20],[52.5,-0.20],[53.5,-0.20],[54.5,-0.20],[55.5,-0.20],[56.5,-0.20],[57.5,-0.40],[58.5,-0.40],[59.5,-0.30],[60.5,-0.30],[61.5,-0.30],[62.5,-0.10],[63.5,-0.30],[64.5,-0.60],[65.5,-0.60],[66.5,-0.40],[67.5,-0.40],[68.5,-0.20],[69.5,0.10],[70.5,0.10],[71.5,-0.10],[72.5,-0.10],[73.5,-0.10],[74.5,-0.40],[75.5,-0.60],[76.5,-0.40],[77.5,-0.60],[78.5,-0.60],[79.5,-0.60],[80.5,-0.40],[81.5,-0.40],[82.5,-0.20],[83.5,-0.40],[84.5,-0.20],[85.5,-0.40],[86.5,-0.40],[87.5,-0.40],[88.5,-0.20],[89.5,-0.20],[90.5,-0.00],[91.5,-0.00],[92.5,-0.00],[93.5,-0.00],[94.5,0.10],[95.5,-0.10],[96.5,-0.10],[97.5,-0.10],[98.5,-0.10],[99.5,-0.20],[100.5,-0.00],[101.5,-0.00],[102.5,-0.00],[103.5,-0.20],[104.5,-0.20],[105.5,-0.20],[106.5,-0.40],[107.5,-0.40],[108.5,-0.40],[109.5,-0.40],[110.5,-0.60],[111.5,-0.40],[112.5,-0.40],[113.5,-0.20],[114.5,-0.20],[115.5,-0.20],[116.5,-0.20],[117.5,-0.20],[118.5,-0.20],[119.5,-0.20],[120.5,-0.20],[121.5,-0.40],[122.5,-0.40],[123.5,-0.40],[124.5,-0.40],[125.5,-0.20],[126.5,-0.00]];dis_datapoints = undefined;trans_datapoints = undefined;sec_helix_datapoints = undefined;sec_strand_datapoints = undefined;acc_exposed_datapoints = undefined;acc_buried_datapoints = undefined;flot_plot_options = []; flot_plot_options[0] = {grid: {borderWidth: {top: 0,right: 0,bottom: 0,left: 0}},legend: {show: false},xaxes: [{show: true,min: 0,max: 200,ticks: [[0.5, '1'], [24.5, '25'], [49.5, '50'], [74.5, '75'], [99.5, '100'], [124.5, '125'], [149.5, '150'], [174.5, '175'], [199.5, '200']],tickLength: -5}],yaxes: [{show: true,ticks: [[0, '0'], [4.5,'hydro-<br>phobic '], [-4.5,'hydro-<br>philic ']],min: -4.5,max: +4.5,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(100,149,237,1)'}},{show: true,ticks: [[0, ''], [1,'positive<br> charge'], [-1,'negative<br> charge']],position: 'right',min: -1,max: 1,font: {size: 12,lineHeight: 14,style: 'italic',weight: 'bold',family: 'sans-serif',variant: 'small-caps',color: 'rgba(255,99,71,1)'}}]};number_of_plots = 1;for ( plot_num = 1 ; plot_num < number_of_plots ; plot_num ++){flot_plot_options[plot_num] = $.extend(true, {} ,flot_plot_options[0]);flot_plot_options[plot_num].xaxes = [{min: plot_num*200,max: (plot_num + 1)*200,ticks: [ [plot_num*200 + 0.5, (plot_num*200 + 1).toString()], [plot_num*200 + 24.5, (plot_num*200 + 25).toString()], [plot_num*200 + 49.5, (plot_num*200 + 50).toString()], [plot_num*200 + 74.5, (plot_num*200 + 75).toString()], [plot_num*200 + 99.5, (plot_num*200 + 100).toString()], [plot_num*200 + 124.5, (plot_num*200 + 125).toString()], [plot_num*200 + 149.5, (plot_num*200 + 150).toString()], [plot_num*200 + 174.5, (plot_num*200 + 175).toString()], [plot_num*200 + 199.5, (plot_num*200 + 200).toString()] ],tickLength: -5}];};try {if( $('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_button').val() =='Show' ){$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_container').css('display','block');$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_button').val('Hide');var description_html = '<div id=\'AutoAnnotator_plot_selectors\'>';description_html = description_html + '<br> <input type=\'checkbox\' id=\'hydrophobicity_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for hydrophobicity (<img src=\'https://static.igem.org/mediawiki/2013/e/e9/TUM13_hydrophobicity_icon.png\' alt=\'blue graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'charge_checkbox\' checked=\'checked\'> Moving average over 5 amino acids for charge (<img src=\'https://static.igem.org/mediawiki/2013/3/3e/TUM13_charge_icon.png\' alt=\'red graph\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'dis_checkbox\' checked=\'checked\'> Predicted disulfid bridges (<img src=\'https://static.igem.org/mediawiki/2013/2/28/TUM13_dis_icon.png\' alt=\'yellow circle\' height=\'10\'></img>) with the number of the bridge in the center';description_html = description_html + '<br> <input type=\'checkbox\' id=\'trans_checkbox\' checked=\'checked\'> Predicted transmembrane helices (<img src=\'https://static.igem.org/mediawiki/2013/7/78/TUM13_trans_icon.png\' alt=\'turquois bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'sec_checkbox\' checked=\'checked\'> Predicted secondary structure: Helices (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_helix_icon.png\' alt=\'violet bars\' height=\'10\'></img>) and beta-strands (<img src=\'https://static.igem.org/mediawiki/2013/b/bf/TUM13_strand_icon.png\' alt=\'yellow bars\' height=\'10\'></img>)';description_html = description_html + '<br> <input type=\'checkbox\' id=\'acc_checkbox\' checked=\'checked\'> Predicted solvent accessability: Exposed (<img src=\'https://static.igem.org/mediawiki/2013/1/16/TUM13_exposed_icon.png\' alt=\'blue bars\' height=\'10\'></img>) and buried (<img src=\'https://static.igem.org/mediawiki/2013/0/0b/TUM13_buried_icon.png\' alt=\'green bars\' height=\'10\'></img>) residues';description_html = description_html + '<br></div>';$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_explanation').html(description_html);plot_according_to_selectors_1381279325962();$('#AutoAnnotator_container_1381279325962 #AutoAnnotator_plot_selectors').find('input').click(plot_according_to_selectors_1381279325962);}else{$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_container').css('display','none');$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_button').val('Show');$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_explanation').html('');}}catch(err){txt='There was an error with the button controlling the visibility of the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';alert(txt);}};function plot_according_to_selectors_1381279325962(){try{var plot_datasets = [[],[]];if($('#AutoAnnotator_container_1381279325962 #hydrophobicity_checkbox').prop('checked') == true){plot_datasets[0] = { color: 'rgba(100,149,237,1)',data: hydrophobicity_datapoints,label: 'Hydrophobicity',lines: { show: true, fill: true, fillColor: 'rgba(100,149,237,0.1)' },yaxis: 1};}if($('#AutoAnnotator_container_1381279325962 #charge_checkbox').prop('checked') == true){plot_datasets[1] = {color: 'rgba(255,99,71,1)',data: charge_datapoints,label: 'Charge',lines: { show: true, fill: true, fillColor: 'rgba(255,99,71,0.1)' },yaxis: 2};}for (plot_num = 0 ; plot_num < number_of_plots ; plot_num ++){$.plot('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder'+ plot_num.toString(), plot_datasets, flot_plot_options[plot_num] );}var screen_width = $('canvas.flot-base').width(); var pos_of_first_tick = 46;var pos_of_last_tick = screen_width - 51;var tick_diff = (screen_width - 97)/199;if($('#AutoAnnotator_container_1381279325962 #dis_checkbox').prop('checked') == true){for ( j = 0 ; j < dis_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][0] - 1)*tick_diff - Math.floor((dis_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder' + Math.floor((dis_datapoints[j][1] - 1)/200) ).append('<div class=\'AutoAnnotator_dis\' style=\'left:' + ((pos_of_first_tick - 8 + (dis_datapoints[j][1] - 1)*tick_diff - Math.floor((dis_datapoints[j][1] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px;\'><b>' + (j+1) + '</b></div>');}}if($('#AutoAnnotator_container_1381279325962 #trans_checkbox').prop('checked') == true){for ( j = 0 ; j < trans_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder' + Math.floor((trans_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_trans\' style=\'width:' + (((trans_datapoints[j][1] - trans_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px ;left:' + ((pos_of_first_tick + (trans_datapoints[j][0] - 1.5)*tick_diff - Math.floor((trans_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if($('#AutoAnnotator_container_1381279325962 #sec_checkbox').prop('checked') == true){for ( j = 0 ; j < sec_helix_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder' + Math.floor((sec_helix_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_helix\' style=\'width:' + (((sec_helix_datapoints[j][1] - sec_helix_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_helix_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_helix_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < sec_strand_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder' + Math.floor((sec_strand_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_sec_strand\' style=\'width:' + (((sec_strand_datapoints[j][1] - sec_strand_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (sec_strand_datapoints[j][0] - 1.5)*tick_diff - Math.floor((sec_strand_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}if($('#AutoAnnotator_container_1381279325962 #acc_checkbox').prop('checked') == true){for ( j = 0 ; j < acc_buried_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder' + Math.floor((acc_buried_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_buried\' style=\'width:' + (((acc_buried_datapoints[j][1] - acc_buried_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_buried_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_buried_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}for ( j = 0 ; j < acc_exposed_datapoints.length ; j++ ){$('#AutoAnnotator_container_1381279325962 #hydrophobicity_charge_placeholder' + Math.floor((acc_exposed_datapoints[j][0] - 1)/200) ).append('<div class=\'AutoAnnotator_acc_exposed\' style=\'width:' + (((acc_exposed_datapoints[j][1] - acc_exposed_datapoints[j][0] + 1)*tick_diff).toFixed(0)).toString() + 'px; left:' + ((pos_of_first_tick + (acc_exposed_datapoints[j][0] - 1.5)*tick_diff - Math.floor((acc_exposed_datapoints[j][0] - 1)/200)*200*tick_diff).toFixed(0)).toString() + 'px\'></div>');}}}catch(err){txt='There was an error while drawing the selected elements for the plot.\n';txt=txt+'The originating error is:\n' + err + '\n\n';throw(txt);}}</script></html> | ||
| + | == References == | ||
| + | <small> | ||
| − | WEIR, J. (2001). Regulation of herpes simplex virus gene expression. ''Gene'' 271: 117-130. | + | [1] WEIR, J. (2001). Regulation of herpes simplex virus gene expression. ''Gene'' 271: 117-130. |
| − | + | [2] GREAVES, R.; O’HARE, P. (1989). Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65.'' J Virol'' 63: 1641-1650. | |
| − | + | [3] TRIEZENBERG, S. J.; KINGSBURY, R. C.; MCKNIGHT, S. L. (1988). Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. ''Genes Dev'' 2: 718-729. | |
| − | + | [4] HIRAI, H.; TANI, T; KIKYO, N. (2010). Structure and functions of powerful transactivators: VP16, MyoD and FoxA. ''Int. J. Dev. Biol.'' 54: 1589-1596. | |
| − | WALKER, R.; GREAVES, R.; O’HARE, P. (1993). Transcriptional activation by the acidic domain and involves additional determinants distinctof Vmw65 requires the integrity of the domain from those necessary for TFIIB binding. ''Molecular and Cellular Biology'' 13: 5233-5244. | + | [5] WALKER, R.; GREAVES, R.; O’HARE, P. (1993). Transcriptional activation by the acidic domain and involves additional determinants distinctof Vmw65 requires the integrity of the domain from those necessary for TFIIB binding. ''Molecular and Cellular Biology'' 13: 5233-5244. |
| − | THOMPSON, R.; PRESTON, C.; SAWTELLl, N. (2009). De Novo Synthesis of VP16 Coordinates the Exit from HSV Latency In Vivo. ''PLoS Pathog'' 5: e1000352. | + | [6] THOMPSON, R.; PRESTON, C.; SAWTELLl, N. (2009). De Novo Synthesis of VP16 Coordinates the Exit from HSV Latency In Vivo. ''PLoS Pathog'' 5: e1000352. |
| + | </small> | ||
| − | |||
| − | |||
| − | |||
| − | + | ==Functional Parameters: Austin_UTexas== | |
| − | + | <html> | |
| + | <body> | ||
<partinfo>BBa_K1150001 parameters</partinfo> | <partinfo>BBa_K1150001 parameters</partinfo> | ||
| − | < | + | <h3><center>Burden Imposed by this Part:</center></h3> |
| + | <figure> | ||
| + | <div class = "center"> | ||
| + | <center><img src = "https://static.igem.org/mediawiki/parts/f/fa/T--Austin_Utexas--no_burden_icon.png" style = "width:160px;height:120px"></center> | ||
| + | </div> | ||
| + | <figcaption><center><b>Burden Value: 2.4 ± 6.4% </b></center></figcaption> | ||
| + | </figure> | ||
| + | <p> Burden is the percent reduction in the growth rate of <i>E. coli</i> cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the | ||
| + | <a href="https://parts.igem.org/Part:BBa_K3174002">BBa_K3174002</a> - <a href="https://parts.igem.org/Part:BBa_K3174007">BBa_K3174007</a> pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the <a href="http://2019.igem.org/Team:Austin_UTexas">2019 Austin_UTexas team</a>.</p> | ||
| + | <p>This functional parameter was added by the <a href="https://2020.igem.org/Team:Austin_UTexas/Contribution">2020 Austin_UTexas team.</a></p> | ||
| + | </body> | ||
| + | </html> | ||
Latest revision as of 18:43, 3 September 2020
VP16
| VP16 | |
|---|---|
| Function | transactivator |
| Use in | Mammalian cells and Herpes simplex virus-1 |
| RFC standard | RFC 25 |
| Backbone | pSB1C3 |
| Submitted by | [http://2013.igem.org/Team:Freiburg Freiburg 2013] |
Virus Protein 16 (VP16) is a transcription factor encoded by the UL48 gene of Herpes simplex virus-1 (HSV-1).
Through complex formation with cellular host factors VP16 can bind to a common regulatory element in the upstream promoter region of immediate-early genes [1]. Due to the transactivating function of VP-16 these genes can then be expressed.
VP16 consists of 490 amino acids with two very important functional domains: a core domain in its central region which is necessary for the indirect DNA binding and a carboxy-terminal transactivation domain [2], [3]; see Fig.1. The transactivation domain (TAD) of VP16 can be fused to a DNA-binding domain (DBD) of another protein in order to gain expression of a desired target gene [1].
Fig. 1: Domain structure of VP16. The different functional domains of VP16 are shown. Taken from Hirai et al., 2010.
Usage and Biology
Transactivation domain (TAD) of VP16
The TAD of VP16 is one of the most efficient TADs. It is widely fused to host transcription factors in order to increase their activity or, next to this, to other transcription factors to study the mechanism of gene regulation [1]. In our case the TAD of VP16 was cloned behind a double mutated Cas9 gene (which encodes for a non-cutting dCas9 protein with specific DNA binding properties) to activate our genes of interest. For more information see BBa_K1150019 and BBa_K1150020.
Biology
VP16 is originally a part of the virus particle from HSV and is released into an animal cell while infection. VP16 first binds via its core domain to the host nuclear protein which then recruits another host nuclear protein, Oct-1, to form a functional protein complex. This complex then binds to its target DNA sequence in the promoter region of immediate-early genes where VP16 activates these genes through interactions between its TAD and many different transcription factors and other proteins, such as histone acetyltransferases; see Fig.2. Through these multiple interactions the transactivation domain of VP16 facilitates the assembly of the pre-initiation complex [5], [1]. Also other TAD containing proteins share this magnitude of binding partners but VP16 is one of the most effective once [1].
Fig. 2: Interaction partners of VP16. Proteins are shown that interact with VP16 in the phase of gene activation. Taken from Hirai et al., 2010
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_K1150001 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGCGCGTACG ... TACGGTGGGACCGGT ORF from nucleotide position -8 to 378 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
References
[1] WEIR, J. (2001). Regulation of herpes simplex virus gene expression. Gene 271: 117-130.
[2] GREAVES, R.; O’HARE, P. (1989). Separation of requirements for protein-DNA complex assembly from those for functional activity in the herpes simplex virus regulatory protein Vmw65. J Virol 63: 1641-1650.
[3] TRIEZENBERG, S. J.; KINGSBURY, R. C.; MCKNIGHT, S. L. (1988). Functional dissection of VP16, the trans-activator of herpes simplex virus immediate early gene expression. Genes Dev 2: 718-729.
[4] HIRAI, H.; TANI, T; KIKYO, N. (2010). Structure and functions of powerful transactivators: VP16, MyoD and FoxA. Int. J. Dev. Biol. 54: 1589-1596.
[5] WALKER, R.; GREAVES, R.; O’HARE, P. (1993). Transcriptional activation by the acidic domain and involves additional determinants distinctof Vmw65 requires the integrity of the domain from those necessary for TFIIB binding. Molecular and Cellular Biology 13: 5233-5244.
[6] THOMPSON, R.; PRESTON, C.; SAWTELLl, N. (2009). De Novo Synthesis of VP16 Coordinates the Exit from HSV Latency In Vivo. PLoS Pathog 5: e1000352.
Functional Parameters: Austin_UTexas
Burden Imposed by this Part:

Burden is the percent reduction in the growth rate of E. coli cells transformed with a plasmid containing this BioBrick (± values are 95% confidence limits). This BioBrick did not exhibit a burden that was significantly greater than zero (i.e., it appears to have little to no impact on growth). Therefore, users can depend on this part to remain stable for many bacterial cell divisions and in large culture volumes. Refer to any one of the BBa_K3174002 - BBa_K3174007 pages for more information on the methods, an explanation of the sources of burden, and other conclusions from a large-scale measurement project conducted by the 2019 Austin_UTexas team.
This functional parameter was added by the 2020 Austin_UTexas team.