Difference between revisions of "Part:BBa K2997002"
| (8 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2997002 short</partinfo> | <partinfo>BBa_K2997002 short</partinfo> | ||
| − | In the human gut microbiome, | + | In the human gut microbiome, <i>Clostridium</i> related species have been reported to have the highest conversion capability of fermenting tyrosine into <i>p</i>-Cresol<sup>[1]</sup>. To target the root of <i>p</i>-Cresol accumulation, reducing the population of <i>Clostridium</i> is needed. We decided to use <i>C. difficile</i> as a model of <i>p</i>-Cresol producing bacteria because it is a popular research target due to its notorious infectious ability. |
| + | Luckily, iGEM NCKU-Tainan 2019 was kindly supported by one of our PIs, Professor Huang, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself to the field of developing a novel therapeutic approach for <i>C. difficile</i> infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) that was proven in his lab to have bactericidal activity against certain strains of <i>Clostridium</i>, including <i>C. difficile</i>. | ||
| + | As proof of the concept that bacteriocin is able to inhibit <i>Clostridium</i> growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with <i>C. difficile</i> R20291 strain. | ||
| + | <html> | ||
| + | <br> | ||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://2019.igem.org/wiki/images/2/2e/T--NCKU_Tainan--spot-on-lawn.png" style="width:35%;"> | ||
| + | </div> | ||
| + | <br> | ||
| + | </html> | ||
| + | Fig. 1. Spot-on-lawn test using 5 μl purified bacteriocin, inhibition zone formation in the middle of the plate can clearly be seen. | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 20: | Line 30: | ||
<partinfo>BBa_K2997002 parameters</partinfo> | <partinfo>BBa_K2997002 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| + | |||
| + | |||
| + | ===Reference=== | ||
| + | [1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and <i>p</i>-Cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. <i>FEMS Microbiology Ecology, 94</i>(9). | ||
Latest revision as of 11:08, 13 December 2019
Bacteriocin short length (sCBM-B)
In the human gut microbiome, Clostridium related species have been reported to have the highest conversion capability of fermenting tyrosine into p-Cresol[1]. To target the root of p-Cresol accumulation, reducing the population of Clostridium is needed. We decided to use C. difficile as a model of p-Cresol producing bacteria because it is a popular research target due to its notorious infectious ability.
Luckily, iGEM NCKU-Tainan 2019 was kindly supported by one of our PIs, Professor Huang, an assistant professor from the Microbiology and Immunology Department, who is currently devoting himself to the field of developing a novel therapeutic approach for C. difficile infection. He kindly provided us with a plasmid containing a bacteriocin gene (CBM-B) that was proven in his lab to have bactericidal activity against certain strains of Clostridium, including C. difficile.
As proof of the concept that bacteriocin is able to inhibit Clostridium growth, we did a spot-on-lawn assay using purified bacteriocin protein provided by advisors to observe the inhibition zone formation. As shown in Fig.1 below, a clear inhibition zone formed in the middle of the BHI plate streak with C. difficile R20291 strain.
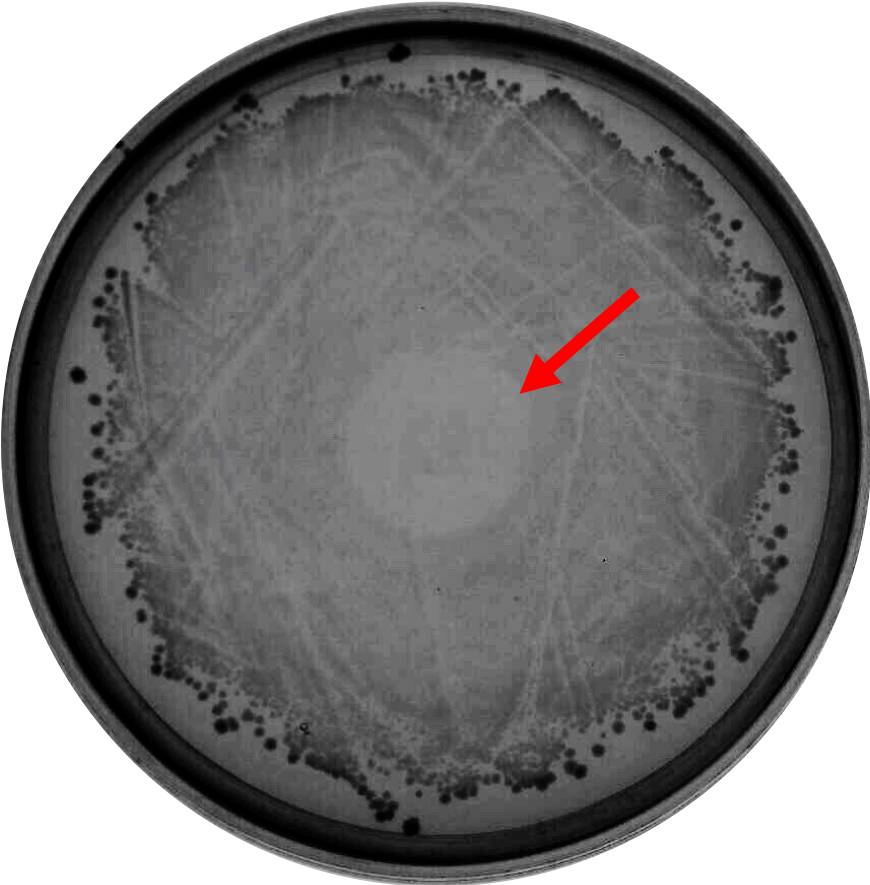
Fig. 1. Spot-on-lawn test using 5 μl purified bacteriocin, inhibition zone formation in the middle of the plate can clearly be seen.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Reference
[1] Saito, Y., Sato, T., Nomoto, K., & Tsuji, H. (2018). Identification of phenol- and p-Cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiology Ecology, 94(9).
