Difference between revisions of "Part:BBa K1438001"
| (5 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| + | |||
<partinfo>BBa_K1438001 short</partinfo> | <partinfo>BBa_K1438001 short</partinfo> | ||
== Optimized bacterioferritin and its properties == | == Optimized bacterioferritin and its properties == | ||
| Line 63: | Line 64: | ||
==================================================================================== | ==================================================================================== | ||
| + | |||
| + | <h2>In silico protein modeling</h2> | ||
| + | |||
<b>Group:</b> CU 2019 | <b>Group:</b> CU 2019 | ||
| − | + | <p>Using Expasy ProtParam tool, theoretical protein extinction coefficient has been identified in addition to several other factors.</p> | |
| − | <p>Using Expasy ProtParam tool, theoretical protein extinction coefficient has been identified in addition to several other factors | + | |
| − | + | <html> | |
| − | <p> | + | <head> |
| − | <p> | + | <style> |
| − | https://2019.igem.org/wiki/images/ | + | |
| + | } | ||
| + | table, th, td { | ||
| + | border: 1px solid black; | ||
| + | border-collapse: collapse; | ||
| + | } | ||
| + | th, td { | ||
| + | padding: 5px; | ||
| + | } | ||
| + | </style> | ||
| + | </head> | ||
| + | <body> | ||
| + | <table style="width:95%"> | ||
| + | <tr> | ||
| + | |||
| + | <th>Extiction Coeficcient</th> | ||
| + | <th>Gravy</th> | ||
| + | <th>PI</th> | ||
| + | <th>Instability Index</th> | ||
| + | <th> Aliphatic Index</th> | ||
| + | <th>kDa</th> | ||
| + | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td>21430</td> | ||
| + | <td>-0.504</td> | ||
| + | <td>4.75</td> | ||
| + | <td>50.08</td> | ||
| + | <td>104.94</td> | ||
| + | <td>17.649</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | <p>Identifying the folding of the protein, and predicting the Gene Ontology, the protein was shown to have metal binding protein capacity for Zinc with a <b>TM-score 0.6052</b>, and Nickel with <b>TM-score0.6078</b>; Whereas in the ligand binding prediction it was predicted to mainly bind copper.</p> | ||
| + | <div style="width:image width 300px; font-size:90%; text-align:center;"><img src="https://2019.igem.org/wiki/images/a/a4/T--CU--bacmodel.png" alt="" width="300px" height="300px" ;" /><br>Fig.1</br></div> | ||
| + | |||
| + | |||
| + | |||
| + | <h2> Bacterioferritin production</h2> | ||
| + | <p>The protein was produced using Ls70 linear expression cell-free kit, under the control of constitutive family member promotor (J23102 ) and strong RBS ( B0034).The range of protein produced using the cell-free expression kit was found to be 2.87-42ug/ml </p> | ||
| + | |||
| + | |||
| + | <h2>Testing the binding affinities of the target protein</h2> | ||
| + | <p>We tested the protein ability to reduce TDS of both sodium chloride and ferric chloride solution the bacterioferritin seemed to have higher affinity for the ferric than sodium salts at room temperature. The results shows that bacterioferritin have decreased TDS of NaCl by 330 ppm compared to 700 ppm in case of FeCl3.[Fig.2].</p> | ||
| + | <div style="width:image width 300px; font-size:90%; text-align:center;"><img src="https://2019.igem.org/wiki/images/thumb/c/cd/T--CU--tds3.png/800px-T--CU--tds3.png" alt="" width="300px" height="300px" ;" /><br>Fig.2</br></div> | ||
| + | |||
| + | |||
| + | <h2>Binding affinities of Bacterioferritin in different temperatures&pHs</h2> | ||
| + | <p>In order to know the best temperature at which the proteins bind, we performed an experiment with three different proteins | ||
| + | putting them in NaCl solution and see how will they work at different temperatures 4,25,37,42,68 Celsius. | ||
| + | The results[Fig.3] showed that ,Bacterioferritin didn’t give any significant result at room temperature,but worked its best at 42 degree reducing total dissolved salts </p> | ||
| + | |||
| + | <div style="width:image width 300px; font-size:90%; text-align:center;"><img src="https://2019.igem.org/wiki/images/thumb/4/47/T--CU--temp.png/800px-T--CU--temp.png" alt="" width="300px" height="300px" ;" /><br>Fig.3</br></div> | ||
| + | |||
| + | |||
| + | <p>While in case of applying solution of same concentration NaCl adjusted to different pHs ranged from 3-9. | ||
| + | Bacterioferritin has highly reduced the value of TDS to around 1200, 1506 ppm respectively. Showing better reduction and binding in acidic media than in Basic media,We added GST to this part and submitted it as BBa_K3144012, were we improved the ability of it to reduce TDS in the sodium chloride solutions.</p> | ||
| + | <div style="width:image width 300px; font-size:90%; text-align:center;"><img src="https://2019.igem.org/wiki/images/thumb/3/3c/T--CU--pH.png/800px-T--CU--pH.png" alt="" width="300px" height="300px" ;" /><br>Fig.4</br></div> | ||
| + | |||
| + | |||
| + | |||
| + | <h2>Testing the bacterioferritin improvement after adding GST Tag</h2> | ||
| − | + | We combined GST and Bacterioferritin in one plasmid under the expression of tac promoter and submitted it as BBa_k3144013 taking into consideration the folding time of the fused protein, then we tested the reduction of the TDS value of NaCl solution at room temperature; as earlier the protein didn't function at that temperature | |
| − | + | From the results we found a great improvement in the ability of the tagged bacterioferritin to reduce the TDS of NaCl solution at room temperature, to around 500 ppm, enabling the fusion protein to behave better in further application of the system [Fig.5] | |
| − | + | </p> | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | https://2019.igem.org/wiki/images/ | + | <div style="width:image width 300px; font-size:90%; text-align:center;"><img src="https://2019.igem.org/wiki/images/thumb/e/ec/T--CU--tag.png/800px-T--CU--tag.png" alt="" width="300px" height="300px" ;" /><br>Fig.5</br></div> |
| − | + | ||
| − | + | ||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- --> | <!-- --> | ||
Latest revision as of 20:45, 12 December 2019
Bacterioferritin (BFR) M52H heme-deletion
Optimized bacterioferritin and its properties
Iron Storaging Protein
Bacterioferritins are the E. coli cells natural iron storage proteins. These hollow nearly spherical protein shells detoxify the cell by sequestering excessive iron and forming Iron(III)hydroxid-oxide particels.
Bacterioferritin is an haem containing bacterial ferritin. Each heme is bound in a pocked formed by the interface between a pair of symmetry-related subunits [1]. However, it was investigated that these heme groups may be involved in the release of iron out of the ferritin iron core by forming an heme-mediated electron transfer to reduce immobilized Fe3+ to more soluble Fe2+.
We isolated this particular bacterioferritin from the probiotic strain E. coli Nissle 1917, which does have more iron aquisition systems than other well established E. coli strains. We conducted a site-directed mutagenesis to produce a haem free bacterioferritin protein [3].
Bacterioferritin as a mediator for magnetism in a cell
Bacterioferritins have been investigated regarding their magnetic character since the 80s. Bfr overexpressing E. coli Nissle 1917 - a strain with a low immunogenity- were previously used in studies as biological MRI contrast agents. [4] Furthermore, the magentic character of iron loading bacterioferritins were studied by Hawkins & Williams concluding that the contamination of the bacterioferritin iron core with phosphate reduces the super paramagnetic properties significantally. [5]
Usage and Biology
Characterization and Function
Group: TAS_Taipei 2019
Author: Allison Kuo, Anna Chang and Yasmin Lin
Summary: We inserted part BBa_1438001 into our composite part BBa_K2921350. Our composite part added a colored protein on the end of BBa_K1438001 in order to see the fusion protein once it is bound to Iron ions.
Characterization
We used SDS-PAGE to check for BacFerr-GS-mRFP expression in E. coli carrying our construct. Bacterial cultures expressing BacFerr-GS-mRFP were grown overnight at 37°C, lysed and run on SDS-PAGE gels. BacFerr-GS-mRFP is approximately 46 kDa, and we observed a strong signal at that size in the BacFerr-GS-mRFP lysate sample, suggesting that BacFerr-GS-mRFP is being expressed in the transformed E. coli.

Functional Assay with Iron
Our construct produces intracellular BacFerr-GS-mRFP proteins expected to increase the cells’ capacity to store iron ions. To test the functionality of this protein, we detected the difference in the iron ion storage capacity of construct-expressing cells and negative-control cells. Thus, our experimental group was cells expressing the BacFerr-GS-mRFP fusion protein. Our negative control group was cells expressing mRFP only. In order to measure cell storage capacity, we incubated cells with the iron ions over time, to allow the iron ions to diffuse in and out of the cell. Theoretically, for our experimental groups, the iron ions would diffuse into the cell and bind to the active site of the intracellular bacterioferritin protein, reducing the amount of iron ions diffusing out of the cell. After 2 hours of incubation, we measured the absorbance of iron ions in the extracellular solution. By the Beer-Lambert law, concentration is directly proportional to absorbance. Thus, for the experimental groups, we expected the extracellular solution to have a lower concentration of iron ions and, thus, a lower absorbance as compared to the negative control. Iron solution was prepared by dissolving in distilled water. To optimize the absorbance measurements in the downstream experiment, the wavelength at the peak absorbance of metal solutions were first determined using a spectrophotometer.

Experimental setup: measuring the peak absorbance of iron solution. FeSO4(H2O)7 was dissolved in distilled water for a 15mM Fe solution. The solution was measured for its absorbance across the full visible light spectrum using a spectrophotometer.
Overnight bacterial cultures were prepared and standardized to an OD600 of 0.7. Then, the cultures were centrifuged and the pellet was resuspended in iron solution. The cell-iron mixtures were gently shaken at room temperature for 2 hours. The cells were then spun down to isolate extracellular solution as the supernatant. The peak absorbance of the iron ions in the supernatant was measured using a spectrophotometer blanked with distilled water.

Experimental setup: measuring extracellular concentrations of protein-cell mixtures. The pelleted bacteria were resuspended in iron solution. After gently shaking the mixture for 2 hours, the absorbance at 776.8 nm of the supernatant was measured using a spectrophotometer. It is expected that the extracellular solution of the experimental group has a lower absorbance than the negative control.
Our results indicate that there are lower absorbance values at the peak absorbance of iron, 776.8 nm, for cells expressing the BacFerr-GS-mRFP fusion protein, as compared to the RFP only negative control. There is a percent difference of -70.7% between the mean absorbance values of the experimental and control group, suggesting a decrease in extracellular iron concentration in the presence of BacFerr-GS-mRFP. This shows that proteins are capable of binding to iron ions, thus increasing the cell’s ability to retain metal ions from their environment.

Bacterioferritin and Bacterioferritin-RFP increase cellular retention of iron ions. After two hours of shaking incubation with 15 mM iron ions, all samples were centrifuged to isolate extracellular solution. At 776.8 nm (the absorbance peak of iron ions), lower absorbance was observed in the extracellular solution of cells expressing BacFerr-GS-mRFP. Cells expressing mRFP only were used as a negative control. Error bars represent standard error. There is a -70.7% percent difference between the mean absorbance values of the experimental and control group, suggesting a decrease in extracellular iron concentration in the presence of BacFerr.
Improving BFRs Iron Storage Properties
We constructed a improved version of this protein by site-directed Mutagenesis (M52H) and added it to the parts registry as BBa_K1438001.
Furthermore we constructed the iGEM Ferritin library which consists of 6 different ferritin protein expression devices, which can be used by future generations of iGEM students to further works with ferritins as nanocages.
All of these parts are featured with an his-tag so purification and invitro studies may be conducted.
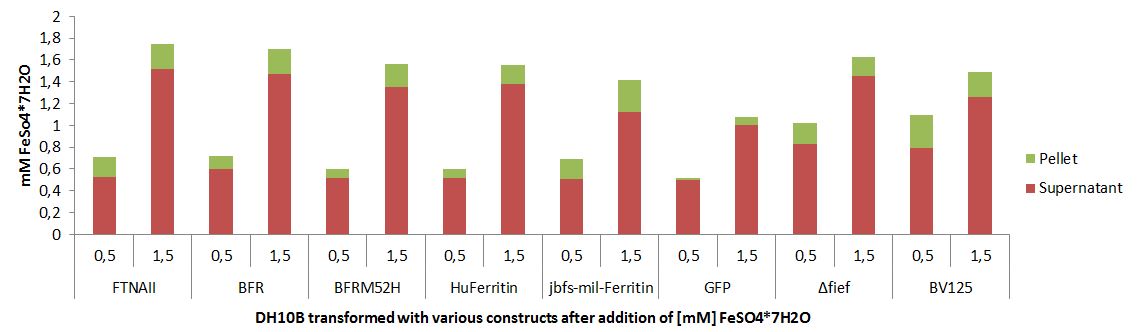
Quantification of Bacterioferritin mediated iron capacity increase
More iron can be stored inside of a cell that is overexpressing bacterioferritin.
========================================================================
In silico protein modeling
Group: CU 2019
Using Expasy ProtParam tool, theoretical protein extinction coefficient has been identified in addition to several other factors.
| Extiction Coeficcient | Gravy | PI | Instability Index | Aliphatic Index | kDa |
|---|---|---|---|---|---|
| 21430 | -0.504 | 4.75 | 50.08 | 104.94 | 17.649 |
Identifying the folding of the protein, and predicting the Gene Ontology, the protein was shown to have metal binding protein capacity for Zinc with a TM-score 0.6052, and Nickel with TM-score0.6078; Whereas in the ligand binding prediction it was predicted to mainly bind copper.

Fig.1
Bacterioferritin production
The protein was produced using Ls70 linear expression cell-free kit, under the control of constitutive family member promotor (J23102 ) and strong RBS ( B0034).The range of protein produced using the cell-free expression kit was found to be 2.87-42ug/ml
Testing the binding affinities of the target protein
We tested the protein ability to reduce TDS of both sodium chloride and ferric chloride solution the bacterioferritin seemed to have higher affinity for the ferric than sodium salts at room temperature. The results shows that bacterioferritin have decreased TDS of NaCl by 330 ppm compared to 700 ppm in case of FeCl3.[Fig.2].

Fig.2
Binding affinities of Bacterioferritin in different temperatures&pHs
In order to know the best temperature at which the proteins bind, we performed an experiment with three different proteins putting them in NaCl solution and see how will they work at different temperatures 4,25,37,42,68 Celsius. The results[Fig.3] showed that ,Bacterioferritin didn’t give any significant result at room temperature,but worked its best at 42 degree reducing total dissolved salts

Fig.3
While in case of applying solution of same concentration NaCl adjusted to different pHs ranged from 3-9. Bacterioferritin has highly reduced the value of TDS to around 1200, 1506 ppm respectively. Showing better reduction and binding in acidic media than in Basic media,We added GST to this part and submitted it as BBa_K3144012, were we improved the ability of it to reduce TDS in the sodium chloride solutions.

Fig.4
Testing the bacterioferritin improvement after adding GST Tag
We combined GST and Bacterioferritin in one plasmid under the expression of tac promoter and submitted it as BBa_k3144013 taking into consideration the folding time of the fused protein, then we tested the reduction of the TDS value of NaCl solution at room temperature; as earlier the protein didn't function at that temperature From the results we found a great improvement in the ability of the tagged bacterioferritin to reduce the TDS of NaCl solution at room temperature, to around 500 ppm, enabling the fusion protein to behave better in further application of the system [Fig.5]
Fig.5