Difference between revisions of "Part:BBa K1819000"
Laisribovski (Talk | contribs) |
(→Characterization by 2019 team XJTU-CHINA) |
||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 10: | Line 10: | ||
</div> | </div> | ||
| − | <img src=https://static.igem.org/mediawiki/2015/5/52/Brasil-USP_NdeIlinkerGFP.png width= "300"> </img src> | + | <center><img src=https://static.igem.org/mediawiki/2015/5/52/Brasil-USP_NdeIlinkerGFP.png width= "300"> </img src><br> |
| − | <p>Schematic representation of BBa_K1819000 insert</p> | + | <p>Figure 1 - Schematic representation of BBa_K1819000 insert</p></center> |
| + | <br> | ||
| + | <h3>Characterization</h3> | ||
| + | <center> | ||
| + | <img src="https://static.igem.org/mediawiki/2015/3/3b/Team-Brasil-USP-LinkerGFP-PSB1C3.png" width= "600"> </img src><br> | ||
| + | <p>Figure 2 - Gel restriction analysis</p></center> | ||
| + | <br> | ||
| − | |||
| − | </ | + | <p>We added this part as an improvement to BBa_E0040, a green fluorescente protein, for protein fusion (see <a href="https://parts.igem.org/Part:BBa_E0040:Experience">Experience page</a>) . |
| + | <br> | ||
| + | This part was characterized using two constitutive promoters from Anderson library (BBa_J23106 and BBa_J23117). See BBa_K1819006 and BBa_K1819007 pages for more information.</p> | ||
| + | |||
| + | |||
| + | </html> | ||
<partinfo>K1819000 SequenceAndFeatures</partinfo> | <partinfo>K1819000 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | ==Characterization by 2019 team XJTU-CHINA== | ||
| + | |||
| + | The characterization of this green fluorescent protein was performed with transcriptional unit BBa_K1819006 which was assembled in vector pSB1C3. In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we assembled with it, we have obtained the emission and excitation spectra in the conditions of our equipment. | ||
| + | |||
| + | Day1: Transform pSB1K3-K1819006 into Escherichia coli DH5α, S17-7 and BL21 respectively. | ||
| + | |||
| + | Day2: Inoculate the transformants in 5-10 mL LB medium with chloramphenicol (25 ug/ml) which was cultured for 12h. | ||
| + | |||
| + | Day3: | ||
| + | |||
| + | 1. Inoculate the recombinant 1% of E.coli into fresh LB medium with chloramphenicol, and culture at 37℃,200 rpm | ||
| + | |||
| + | 2. Measure OD600 and GFP fluorescent intensity (RFU) of the bacteria every 2 hours using 200 ul of the culture broth with six parallel groups. | ||
| + | |||
| + | 3. We will test GFP emission scan curve from 495 nm to 580 nm when the excitation wavelength was fixed at 475 nm, GFP excitation scan curve from 430 nm to 520 nm with the fixed emission at 545 nm. | ||
| + | |||
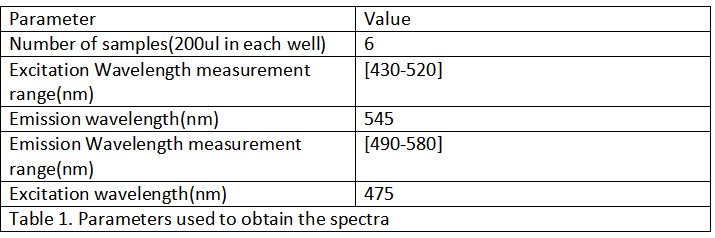
| + | By using this protocol with the parameters of Table 1, Figure 1 was obtained. | ||
| + | |||
| + | <div>[[File:T--XJTU-CHINA--TABLE1.png |700px|thumb|center|]]</div> | ||
| + | |||
| + | |||
| + | |||
| + | <div>[[File:T--XJTU-CHINA--EX.png |700px|thumb|center|<b>Figure 1:</b> GFP emission and excitation spectra at different time point for three strains (Ex fixed). | ||
| + | (a) GFP emission and excitation spectra at different time point for DH5α (Ex fixed) | ||
| + | (b) GFP emission and excitation spectra at different time point for S17-1 (Ex fixed) | ||
| + | (c) GFP emission and excitation spectra at different time point for BL21 (Ex fixed)]]</div> | ||
| + | |||
| + | From Fig 1 it is clear to show that the intensity of GFP was gradually increased during the 60h cultivation in all three strains, and the wavelength with highest emission located around 515 nm when the excitation wavelength was fixed at 475 nm. There was no significant difference in the spectra profile of GFP between three E. coli strains, while the intensity of GFP varied in three strains, with the highest intensity in S17-1 at 3.9x107, and lowest in BL21 at 2.0x107. | ||
| + | |||
| + | <div>[[File:T--XJTU-CHINA--OD.png |700px|thumb|center|<b>Figure 2:</b> Figure 2. GFP emission and excitation spectra at different time point for three strains (Em fixed). (a) GFP emission and excitation spectra at different time point for DH5α (Em fixed).(b) GFP emission and excitation spectra at different time point for S17-1 (Em fixed).(c) GFP emission and excitation spectra at different time point for BL21 (Em fixed).]]</div> | ||
| + | |||
| + | When the scanning of excitation wavelength was performed with emission wavelength was fixed at 545nm, similar results were achieved. The intensity of GFP was stably increased and the wavelength with highest excitation was around 505 nm when the emission wavelength was fixed in all three stains. And the three strains shared quite similar spectra profile of GFP, but the intensity of GFP differed, with the highest intensity in S17-1 at 2.6x107, and lowest in BL21 at 1.3x107. | ||
| + | |||
| + | All in all, our validation of the emission and excitation spectra of this GFP in three different recombinant E.coli, provides us the precise wavelength for testing of our parts, as well as demonstrated that the host will not affect the spectra profile of GFP but influence the intensity. From the results above, we can see that the maximum excitation wavelength of the GFP in this part is around 505nm, and the maximum emission wavelength is around 515 nm. | ||
Latest revision as of 03:53, 22 October 2019
Designed by Brasil-USP team, BBa_K1819000 is a GFP coding sequence with an N-terminal linker, which is 5’-flanked by NdeI restriction site.
It allows to characterize protein expression and secretion and also may serve to other IGEM teams that would like to express a chimera C-terminally attached to GFP, which exhibits bright green fluorescence when exposed to light in the blue to ultraviolet range.
We also added GGGS amino acid twice as a spacer to avoid interaction between two consecutive proteins, allowing proper folding. This strategy was already applied to BBa_K1489002 part.

Figure 1 - Schematic representation of BBa_K1819000 insert
Characterization

Figure 2 - Gel restriction analysis
We added this part as an improvement to BBa_E0040, a green fluorescente protein, for protein fusion (see Experience page) .
This part was characterized using two constitutive promoters from Anderson library (BBa_J23106 and BBa_J23117). See BBa_K1819006 and BBa_K1819007 pages for more information.
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 674
Characterization by 2019 team XJTU-CHINA
The characterization of this green fluorescent protein was performed with transcriptional unit BBa_K1819006 which was assembled in vector pSB1C3. In order to carry out a correct characterization of the protein and to be able to use it to make measurements of the different transcriptional units that we assembled with it, we have obtained the emission and excitation spectra in the conditions of our equipment.
Day1: Transform pSB1K3-K1819006 into Escherichia coli DH5α, S17-7 and BL21 respectively.
Day2: Inoculate the transformants in 5-10 mL LB medium with chloramphenicol (25 ug/ml) which was cultured for 12h.
Day3:
1. Inoculate the recombinant 1% of E.coli into fresh LB medium with chloramphenicol, and culture at 37℃,200 rpm
2. Measure OD600 and GFP fluorescent intensity (RFU) of the bacteria every 2 hours using 200 ul of the culture broth with six parallel groups.
3. We will test GFP emission scan curve from 495 nm to 580 nm when the excitation wavelength was fixed at 475 nm, GFP excitation scan curve from 430 nm to 520 nm with the fixed emission at 545 nm.
By using this protocol with the parameters of Table 1, Figure 1 was obtained.
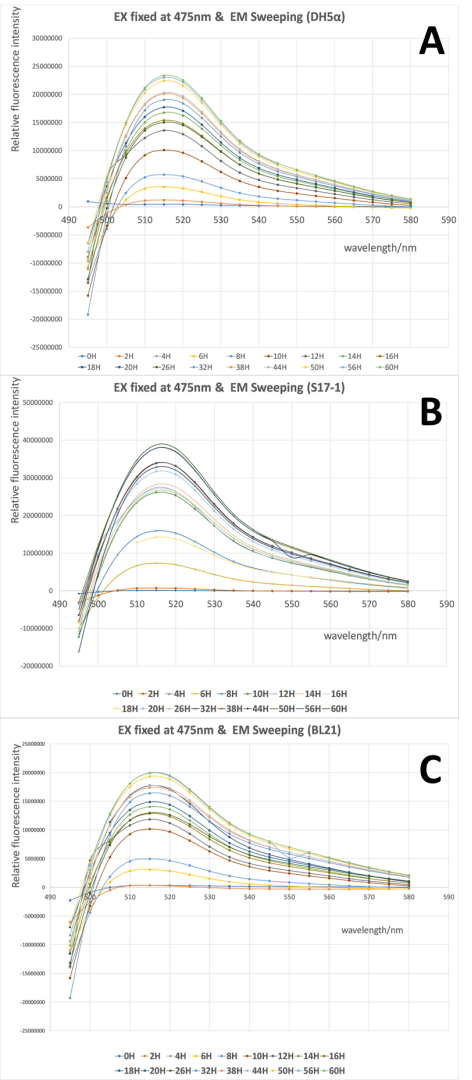
From Fig 1 it is clear to show that the intensity of GFP was gradually increased during the 60h cultivation in all three strains, and the wavelength with highest emission located around 515 nm when the excitation wavelength was fixed at 475 nm. There was no significant difference in the spectra profile of GFP between three E. coli strains, while the intensity of GFP varied in three strains, with the highest intensity in S17-1 at 3.9x107, and lowest in BL21 at 2.0x107.

When the scanning of excitation wavelength was performed with emission wavelength was fixed at 545nm, similar results were achieved. The intensity of GFP was stably increased and the wavelength with highest excitation was around 505 nm when the emission wavelength was fixed in all three stains. And the three strains shared quite similar spectra profile of GFP, but the intensity of GFP differed, with the highest intensity in S17-1 at 2.6x107, and lowest in BL21 at 1.3x107.
All in all, our validation of the emission and excitation spectra of this GFP in three different recombinant E.coli, provides us the precise wavelength for testing of our parts, as well as demonstrated that the host will not affect the spectra profile of GFP but influence the intensity. From the results above, we can see that the maximum excitation wavelength of the GFP in this part is around 505nm, and the maximum emission wavelength is around 515 nm.