Difference between revisions of "Part:BBa K2980009"
| (One intermediate revision by the same user not shown) | |||
| Line 45: | Line 45: | ||
| − | Finally, we verified the regulation ability of our system by downstream reactions, catalyzed by Renilla luciferase and catechol dioxygenase. | + | Finally, we verified the regulation ability of our system by downstream reactions, catalyzed by Renilla luciferase and catechol dioxygenase. By introducing this part in our system, those two reactions could be accelerated by our system. See PART III of [[https://2019.igem.org/Team:Tsinghua/Demonstrate]] for more details about this part. |
Latest revision as of 03:51, 22 October 2019
CIB1-GCN(4)-mEGFP-FUSLCD
This fusion protein can induce consistant phase-separation in cell once well expressed. Under blue light stimulation, Cry2-mCherry (Part:BBa K2980006) will be recruited into its phase.
Phase separation
We cloned the Low Complexity Domain (LCD) of the Fused in Sarcoma (FUS) protein (FUSLCD), which was reported to phase separate spontaneously at high concentrations in vitro due to their multiple weakly adhesive sequence elements. We fused mEGFP (for visualization) and GCN(4) (for spontaneous formation of tetramers) with FUSLCD and expressed GFP-GCN(4)-FUSLCD in E.coli (element 1). Spherical green droplets indicating phase separation were observed (Figure 1A) at two ends of E.coli. The fast Fluorescent Recovery After Photobleaching (FRAP) (Figure 1B) indicated the high fluidity of puncta and validated the feasibility and effectiveness of GCN(4)-FUSLCD phase separation system.
Light stimulation
To achieve the response to light stimulation, two types of light-sensitive protein, CIB1(Part:BBa K2980002) and Cry2(Part:BBa K2980000), are fused to our phase separation elements and application elements, respectively. When exposed to 488nm laser, CIB1 and Cry2 will bind to each other. Since CIB1 is primarily enriched in the compartment formed by phase separation elements (Part:BBa K2980009), Cry2 would be recruited to phase, as the 'switch' in our PhASE turns on. Therefore, the distribution of enzyme or other proteins fused to Cry2 would be altered by light stimulation. (Figure 2A)
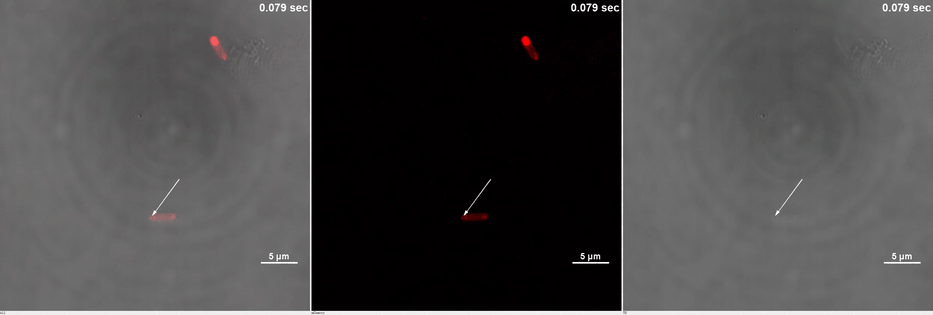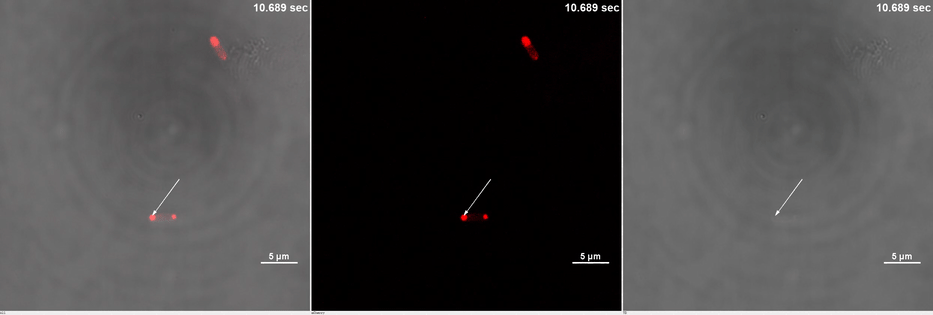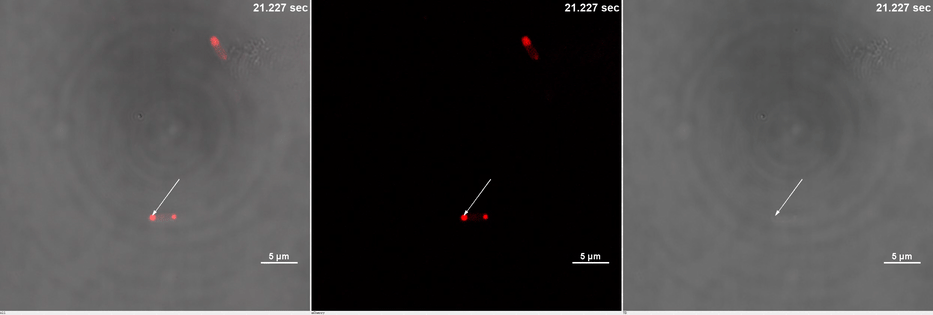
E. coli transformed with CIB1-GCN(4)-mEGFP-FUS (Part:BBa K2980009) and Cry2-mCherry are placed under confocal microscope. Only mCherry channel and TD channel are shown in the gif here (Media 1), since 488 nm laser, which is used to excite mEGFP, can also lead to the binding of CIB1 and Cry2. At 0 second, Cry2-mCherry is almost smear in the cell. (Figure 2B)
Yet, after stimulation, it is recruited to the ends of the cell and form two sphere-like droplets. The screen shots below show the distribution of mCherry before and after 488 nm laser stimulation. (Figure 2C) In order to reflect the recruitment of Cry2-mCherry into phase, we use the ratio of light intensity in phase to the rest of the cell as a standard. (Figure 2D) As presented in the plot, this ratio quickly increases after stimulation and can stay at a rather static level for a long time. (Figure 2E)
-
 Figure 2D. This image shows the selected areas for quantifying phase separation. Among the selected areas, “B” represents for background; “R” represents for reference; “S” represents for stimulation. After comparing light intensity with background and reference bacterium, we can demonstrate the relative change of light intensity in different area of the target bacterium.
Figure 2D. This image shows the selected areas for quantifying phase separation. Among the selected areas, “B” represents for background; “R” represents for reference; “S” represents for stimulation. After comparing light intensity with background and reference bacterium, we can demonstrate the relative change of light intensity in different area of the target bacterium. -
 Figure 2E. Quantified result is shown in this plot. We use the ratio of light intensity in phase to cytosol to reflect the distribution difference before and after stimulation. The result turns out that the this ratio is significantly increased after stimulation at 0 second, which means most elements 2 get into phase after stimulation.
Figure 2E. Quantified result is shown in this plot. We use the ratio of light intensity in phase to cytosol to reflect the distribution difference before and after stimulation. The result turns out that the this ratio is significantly increased after stimulation at 0 second, which means most elements 2 get into phase after stimulation.
What is more, we validated the ability of reversible manner of our switch. Since bacteria on a single slide cannot be observed for a long time, we acquired sample from a culture dish at different time point. After incubated bacteria in dark for a few hours, we got the first sample, which shows smear distribution in the cell. (Figure 3A)
Then, we exposed the whole dish to 488 nm laser for about 20 seconds. Immediately after that, we acquired another sample, with Cry2-mCherry aggregates at the end of the cell. (Figure 3B)
After that, the dish was placed in dark again for half an hour. Next, another sample was acquired, in which Cry2-mCherry reversed to smear state. Finally, we tried to turn on the switch again, which worked as well as previous attempts. (Figure 3C,3D)
Finally, we verified the regulation ability of our system by downstream reactions, catalyzed by Renilla luciferase and catechol dioxygenase. By introducing this part in our system, those two reactions could be accelerated by our system. See PART III of [[1]] for more details about this part.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 112
- 1000COMPATIBLE WITH RFC[1000]