Difference between revisions of "Part:BBa K3304000"
PlatonMega (Talk | contribs) |
|||
| (17 intermediate revisions by 2 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3304000 short</partinfo> | <partinfo>BBa_K3304000 short</partinfo> | ||
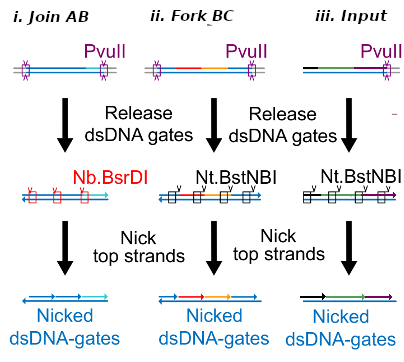
| − | This is the Input Gate for our Chemical Reaction Network. The oligo input is DTG | + | This is the Input Gate for our Chemical Reaction Network. The oligo input is DTG [[Part:BBa_K3304005]] with initiates the strand displacement cascades. Other auxiliary strands for this gate are BTD [[Part:BBa_K3304006]] and NTB [[Part:BBa_K3304007]] that conclude the reactions and facilitate their trajectory to the desired direction. The output of this gate is TBB [[Part:BBa_K3304006]] which constitutes the input for the downstream Join Gate [[Part:BBa_K3304001]]. The part comes with two PvuII cut sites and three Nt.BstNBI nicking sites that facilitate the extraction of the gate from plasmid DNA and prepare the double-stranded block for strand displacement reactions. |
| + | The gate incorporates the consensus binding site for the p65 homodimer in the TDD domain. There the TF can be bound and inhibit the initiation of strand displacement reactions that feed forward the circuit producing the input. | ||
| + | Specific primers that were designed to insert site directed mutations to the Tfs binding site can be deployed in order to alter the sequence of the binding site. They can be used to examine the effect of various SNPs on the TFs binding affinity.The universal forward primer [[Part:BBa_K3304104]] can be used in combination with [[Part:BBa_K3304101]] to change the Thymine in the 8th place to Adenine, with [[Part:BBa_K3304102]] to change the Cytocine in position 10 to Thymine and [[Part:BBa_K3304001]] to change the Thymine in position 6 to Adenine. The gate is incorporated into plasmid psb1c3 through the golden gate assembly method from our modified AmilcP construct [[Part:BBa_K3304016]] | ||
| + | https://2019.igem.org/wiki/images/c/ca/T--Thessaloniki--animation-step1.gif | ||
| + | https://2019.igem.org/wiki/images/0/02/T--Thessaloniki--animation-step2.gif | ||
| + | https://2019.igem.org/wiki/images/3/38/T--Thessaloniki--animation-step3.gif | ||
| + | Animation describing the subsequent strand displacement Cascade steps happening in the Input Gate when no TF is bound | ||
| + | |||
| + | https://2019.igem.org/wiki/images/0/07/T--Thessaloniki--design-figure4.png | ||
| + | |||
| + | https://2019.igem.org/wiki/images/6/62/T--Thessaloniki--results-f2.png | ||
| + | Circuit behavior with gradient concentrations of Input Gate. Furthermore the difference in circuit leakage between plasmid derived and synthetic Gates is shown indicating the superiority of plasmid DNA for the production of DSD Gates | ||
| + | https://2019.igem.org/wiki/images/6/61/T--Thessaloniki--results-f4.png | ||
| + | Fluorescent measurements of circuit behavior after the binding of the p65 homodimer to the consensus sequence. Kinetics are compared in regards to Input Gate concentration titrations | ||
| + | https://2019.igem.org/wiki/images/0/0c/T--Thessaloniki--results-f6.png | ||
| + | Kinetics behavior of the circuit with the mutated binding site for the p65 Transcription Factor. It is apparent that SNP that replaces the cytosine of the 10th nucleotide of the binding site to Thymine retards the ability of the protein to bind Dna completely. The rest of the site directed mutations have an in between behavior expressing gradient binding affinities depending on the nucleotides position | ||
| + | T--Thessaloniki--wiki1.png | ||
| + | Native PAGE gel for the purification of synthetic gates | ||
| + | https://2019.igem.org/wiki/images/b/b0/T--Thessaloniki--bbak33040105.png | ||
| + | Agaroze gel electrophoresis of the plasmid derived gates and the directed mutagenesis derived SNPsafter a standard PCR amplification protocol | ||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 03:48, 22 October 2019
Input Gate with NF-κB p65 consensus sequence
This is the Input Gate for our Chemical Reaction Network. The oligo input is DTG Part:BBa_K3304005 with initiates the strand displacement cascades. Other auxiliary strands for this gate are BTD Part:BBa_K3304006 and NTB Part:BBa_K3304007 that conclude the reactions and facilitate their trajectory to the desired direction. The output of this gate is TBB Part:BBa_K3304006 which constitutes the input for the downstream Join Gate Part:BBa_K3304001. The part comes with two PvuII cut sites and three Nt.BstNBI nicking sites that facilitate the extraction of the gate from plasmid DNA and prepare the double-stranded block for strand displacement reactions. The gate incorporates the consensus binding site for the p65 homodimer in the TDD domain. There the TF can be bound and inhibit the initiation of strand displacement reactions that feed forward the circuit producing the input. Specific primers that were designed to insert site directed mutations to the Tfs binding site can be deployed in order to alter the sequence of the binding site. They can be used to examine the effect of various SNPs on the TFs binding affinity.The universal forward primer Part:BBa_K3304104 can be used in combination with Part:BBa_K3304101 to change the Thymine in the 8th place to Adenine, with Part:BBa_K3304102 to change the Cytocine in position 10 to Thymine and Part:BBa_K3304001 to change the Thymine in position 6 to Adenine. The gate is incorporated into plasmid psb1c3 through the golden gate assembly method from our modified AmilcP construct Part:BBa_K3304016
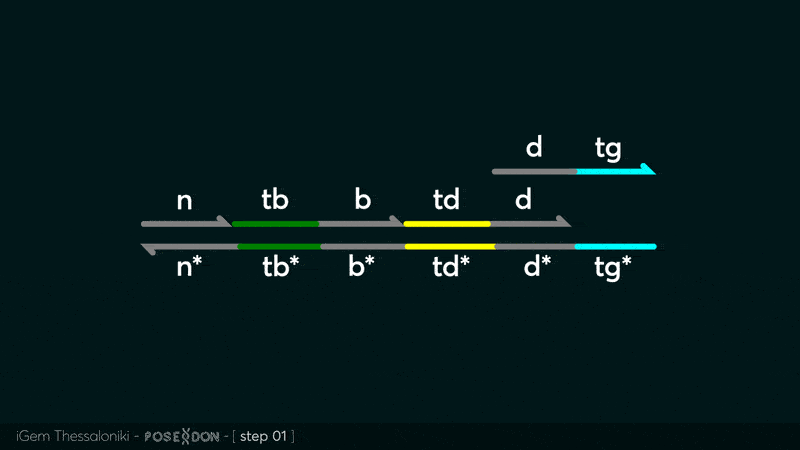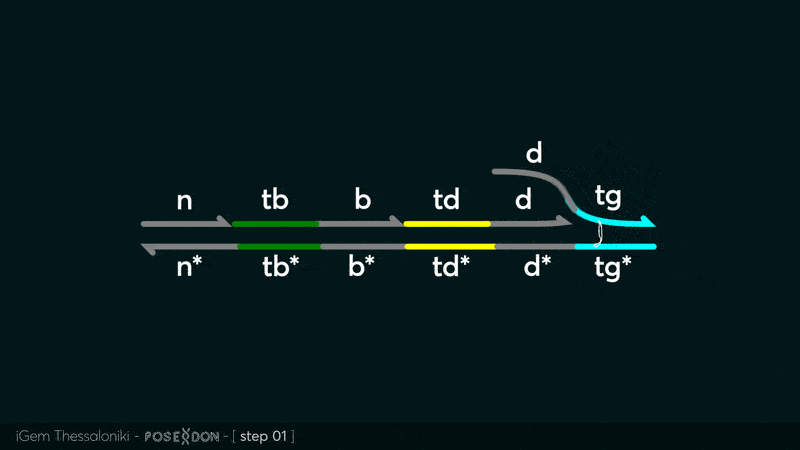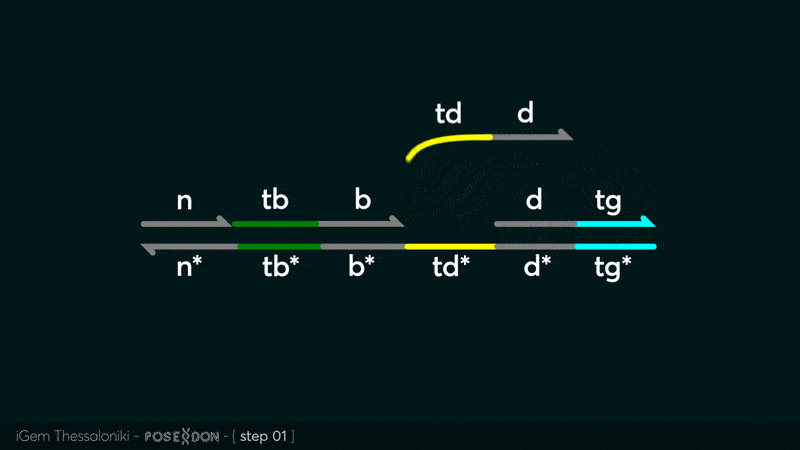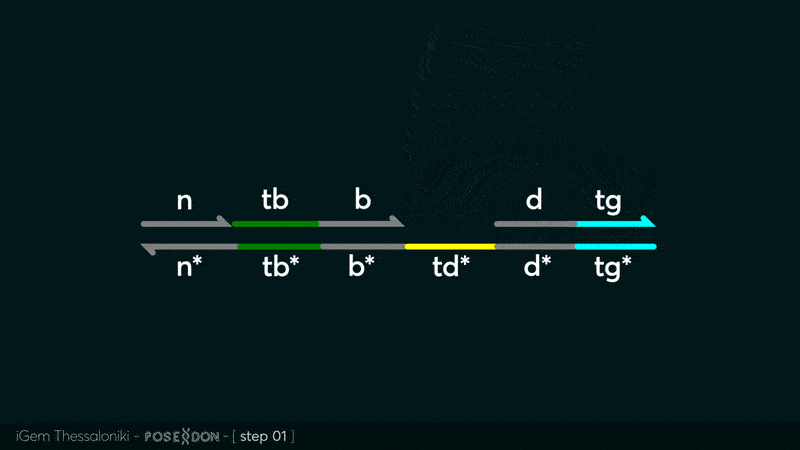
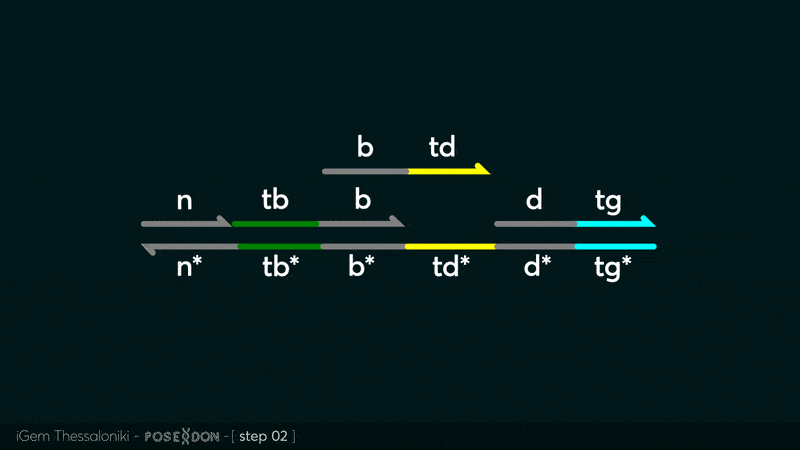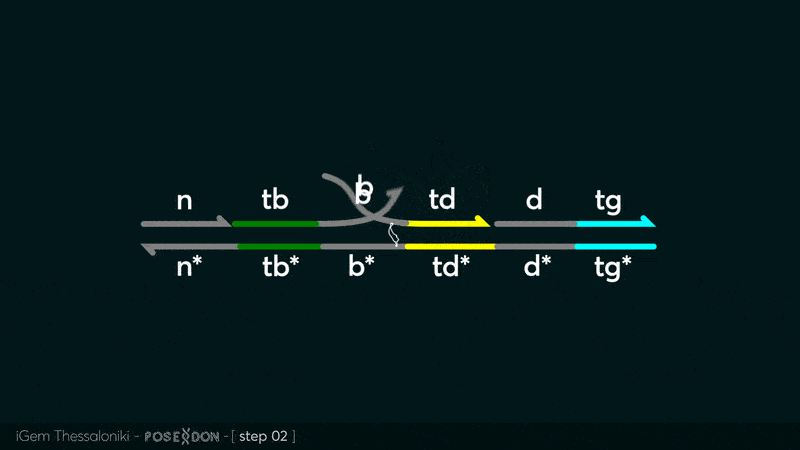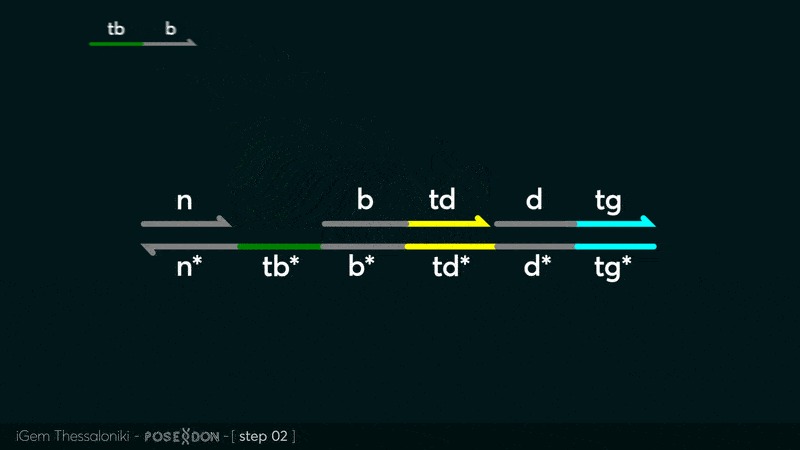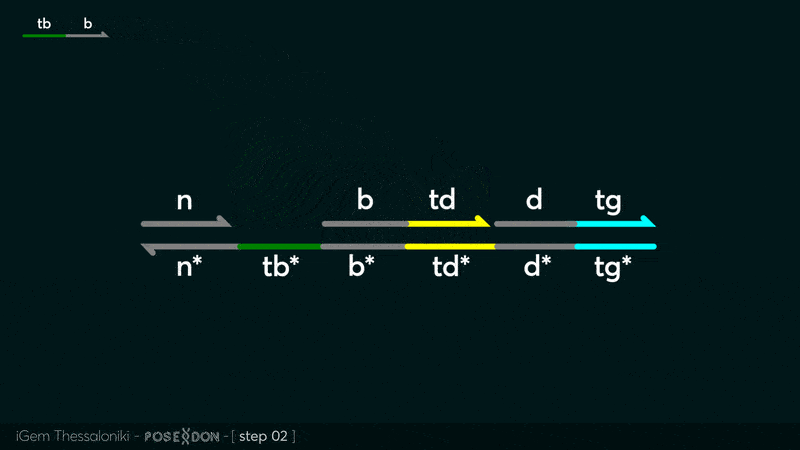
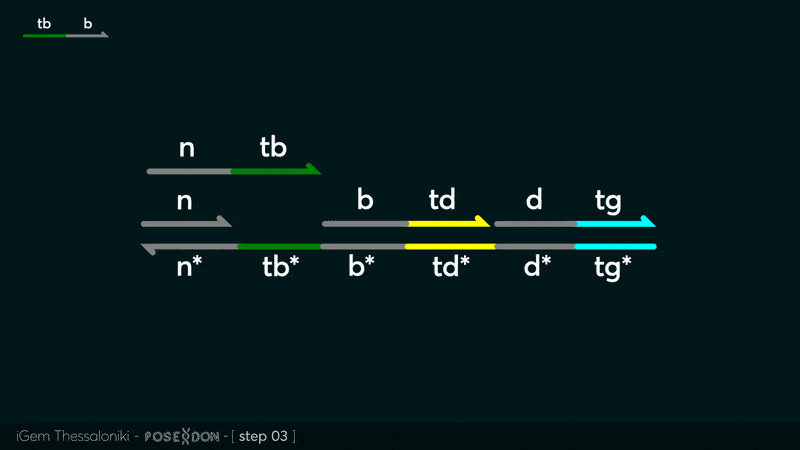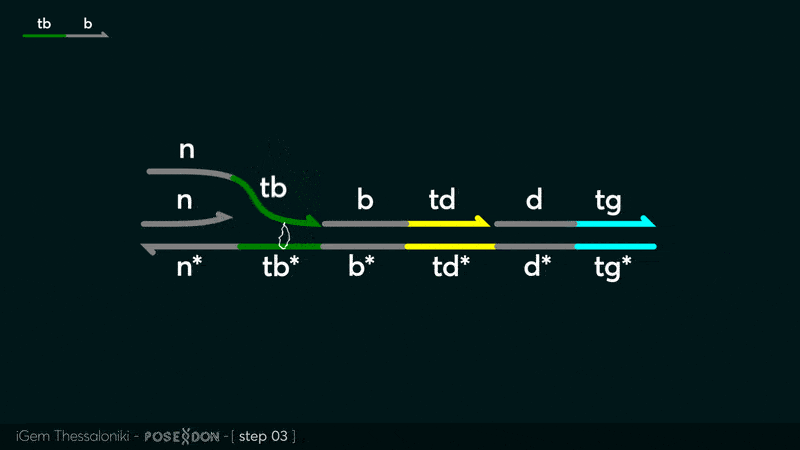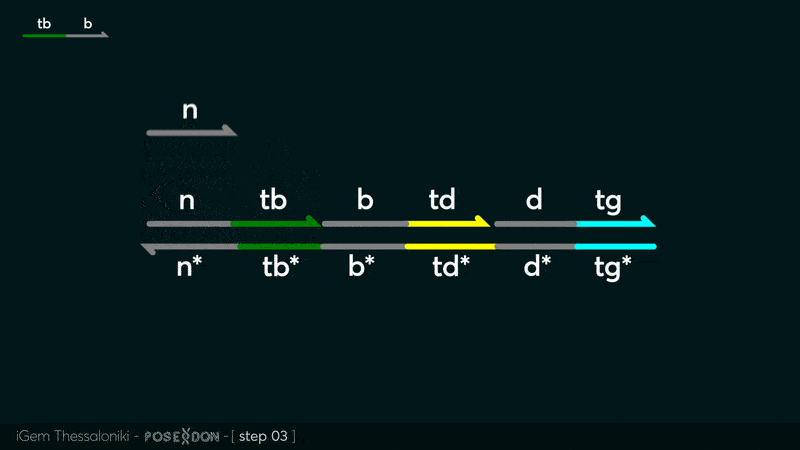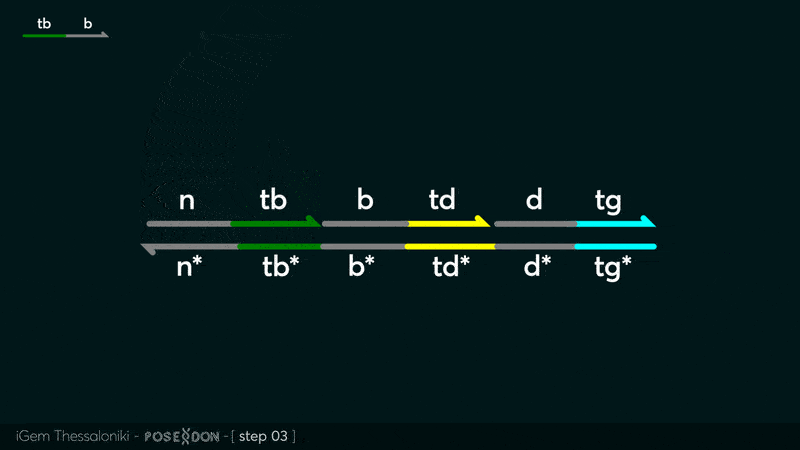 Animation describing the subsequent strand displacement Cascade steps happening in the Input Gate when no TF is bound
Animation describing the subsequent strand displacement Cascade steps happening in the Input Gate when no TF is bound
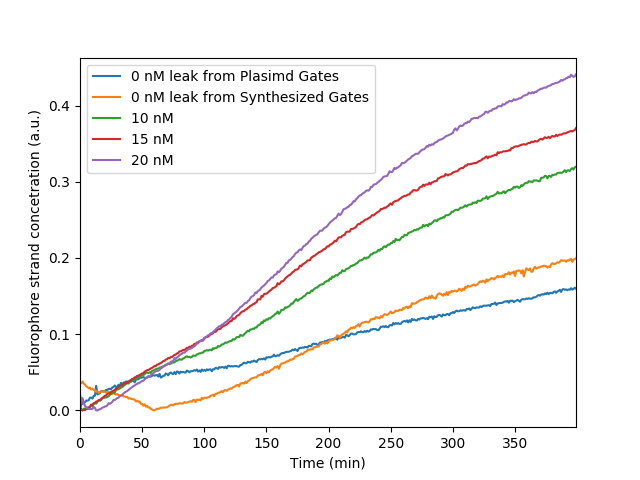 Circuit behavior with gradient concentrations of Input Gate. Furthermore the difference in circuit leakage between plasmid derived and synthetic Gates is shown indicating the superiority of plasmid DNA for the production of DSD Gates
Circuit behavior with gradient concentrations of Input Gate. Furthermore the difference in circuit leakage between plasmid derived and synthetic Gates is shown indicating the superiority of plasmid DNA for the production of DSD Gates
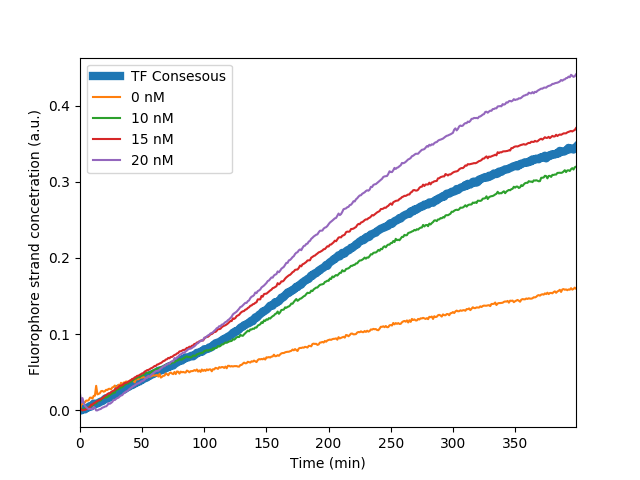 Fluorescent measurements of circuit behavior after the binding of the p65 homodimer to the consensus sequence. Kinetics are compared in regards to Input Gate concentration titrations
Fluorescent measurements of circuit behavior after the binding of the p65 homodimer to the consensus sequence. Kinetics are compared in regards to Input Gate concentration titrations
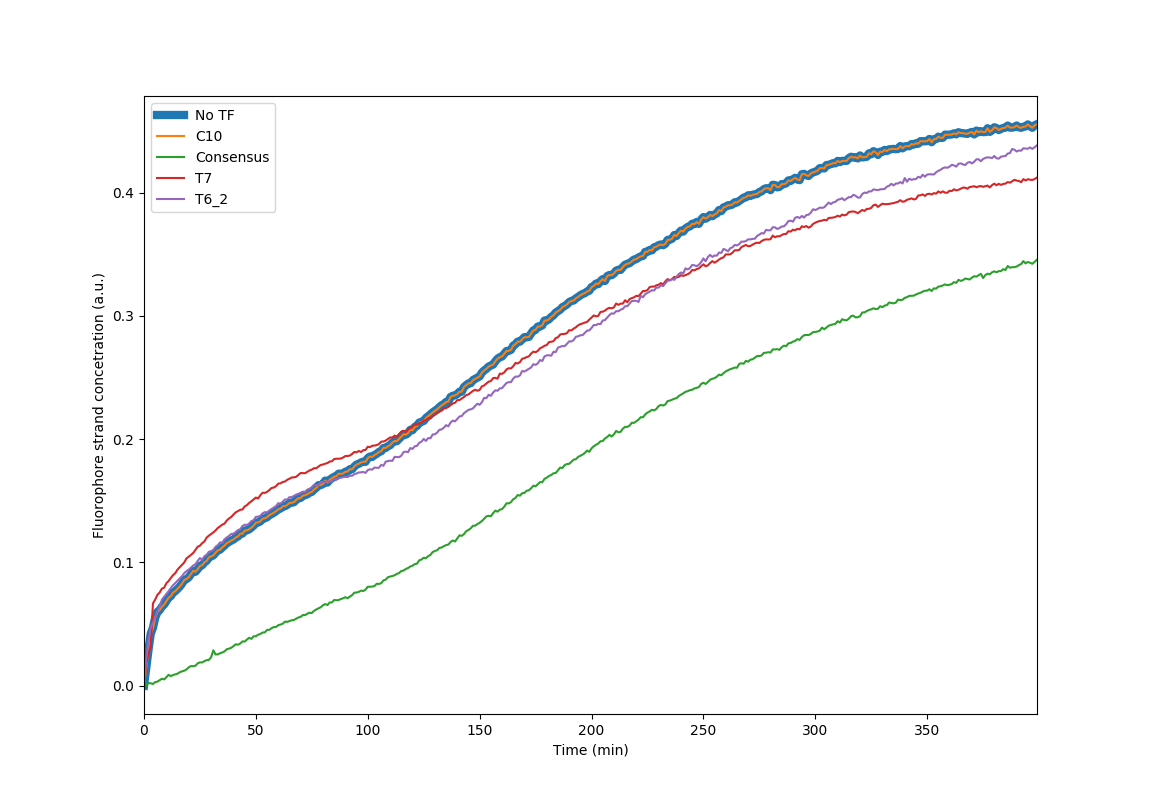 Kinetics behavior of the circuit with the mutated binding site for the p65 Transcription Factor. It is apparent that SNP that replaces the cytosine of the 10th nucleotide of the binding site to Thymine retards the ability of the protein to bind Dna completely. The rest of the site directed mutations have an in between behavior expressing gradient binding affinities depending on the nucleotides position
T--Thessaloniki--wiki1.png
Native PAGE gel for the purification of synthetic gates
Kinetics behavior of the circuit with the mutated binding site for the p65 Transcription Factor. It is apparent that SNP that replaces the cytosine of the 10th nucleotide of the binding site to Thymine retards the ability of the protein to bind Dna completely. The rest of the site directed mutations have an in between behavior expressing gradient binding affinities depending on the nucleotides position
T--Thessaloniki--wiki1.png
Native PAGE gel for the purification of synthetic gates
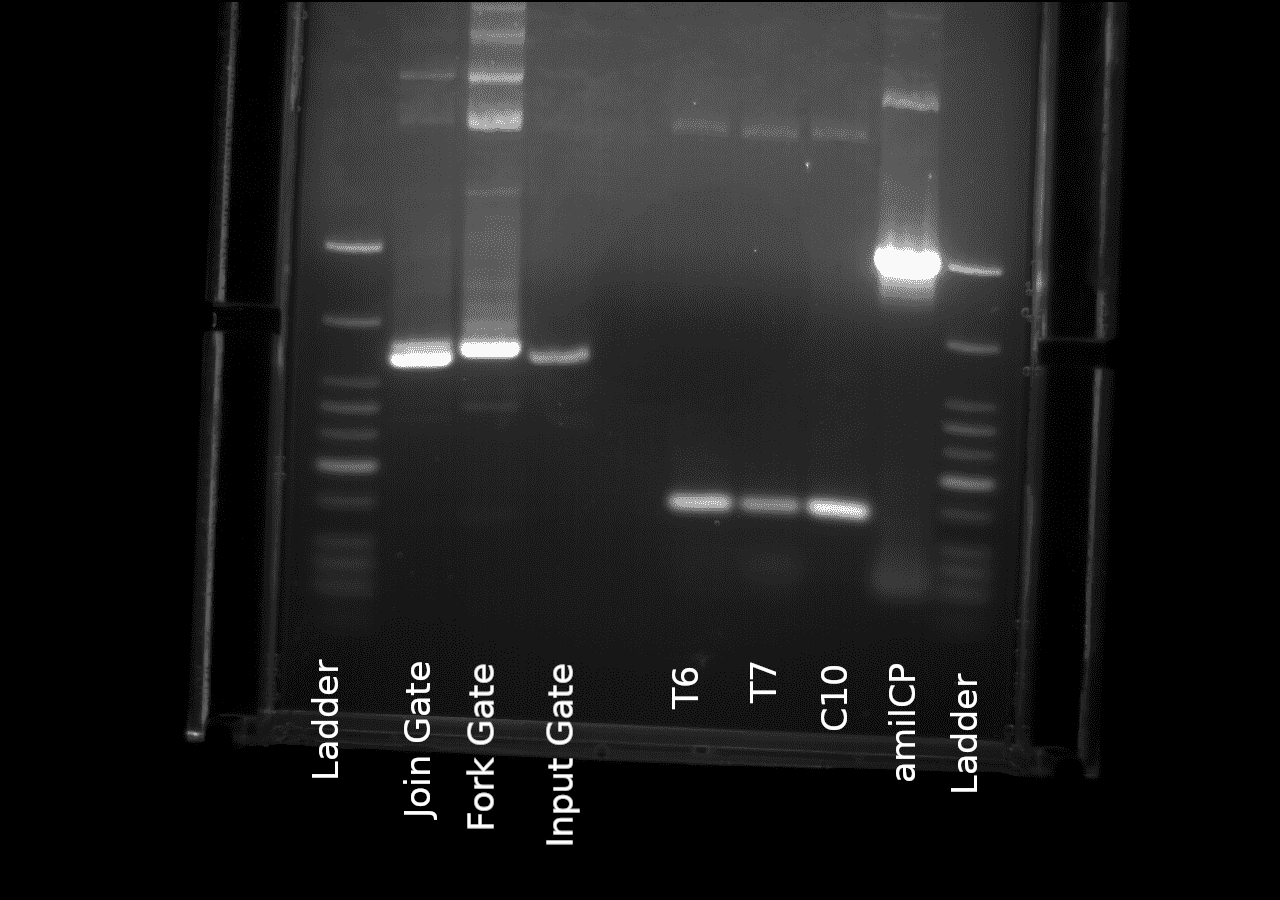 Agaroze gel electrophoresis of the plasmid derived gates and the directed mutagenesis derived SNPsafter a standard PCR amplification protocol
Sequence and Features
Agaroze gel electrophoresis of the plasmid derived gates and the directed mutagenesis derived SNPsafter a standard PCR amplification protocol
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
