Difference between revisions of "Part:BBa K1499500"
| (7 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
<partinfo>BBa_K1499500 short</partinfo> | <partinfo>BBa_K1499500 short</partinfo> | ||
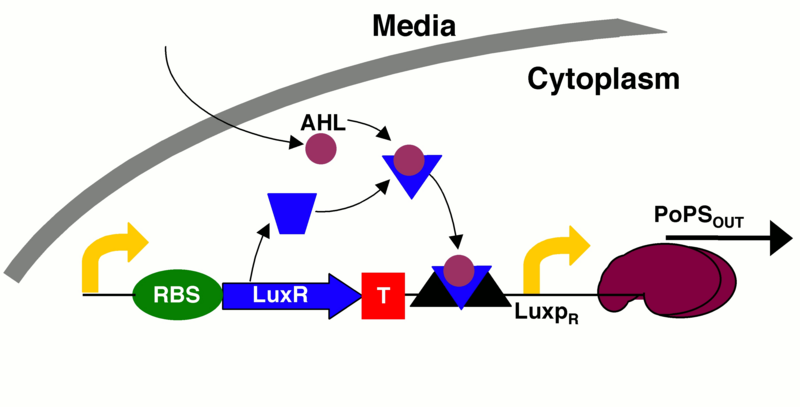
| − | This part is the combination of two parts, BBa_I13202 and BBa_T9002. The first part (BBa_I13202) produces a luxI enzyme which produces an AHL (3OC6HSL). This part is under the control of a lac repressible promoter. The second part (BBa_T9002) produces luxR, which combines with the AHL to activate the luxPR promoter that controls expression of GFP (see image below | + | This part is the combination of two parts, BBa_I13202 and BBa_T9002. The first part (BBa_I13202) produces a luxI enzyme which produces an AHL (3OC6HSL). This part is under the control of a lac repressible promoter. The second part (BBa_T9002) produces luxR, which combines with the AHL to activate the luxPR promoter that controls expression of GFP (see image below [1]). |
[[File:luxoperon-cambridge2010.jpg]] | [[File:luxoperon-cambridge2010.jpg]] | ||
| − | |||
| Line 13: | Line 12: | ||
| + | == Usage and Biology == | ||
| + | |||
| + | |||
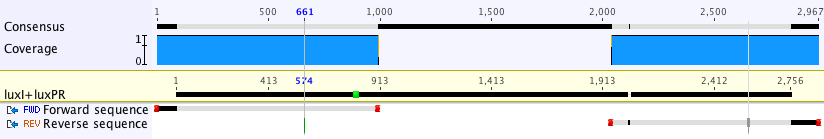
| + | After ligating this construct together and into the BioBrick backbone, we transformed it into the NEB 5-alpha strain of E. coli. The colonies that fluoresced were most likely to have been successfully transformed, and thus these were sent off for sequencing. The sequencing data showed that our construct was correct (see below), so we were able to submit for BioBricking. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:lux sequencing data.jpg]] | ||
| + | |||
| + | |||
| + | |||
| + | Although our sequencing data was good, we realized that we had not induced the lac-repressible promoter, and thus the colonies should not have been fluorescing yet. After doing more research, we realized that the strain of E. coli we were using was not lactose deficient, thus the quorum sensing cascade was being activated by the natural presence of lactose. We ordered a lacI^q strain of E. coli, which are lac deficient, so that we could have complete control of GFP expression. However, when we performed the transformation of the construct into this strain, all of the colonies still fluoresced. | ||
| + | |||
| + | After doing further research, we discovered that the luxPR promoter (BBa_R0062) [2], will induce backward transcription if the luxR protein is present but the AHL molecule is not. In the lac deficient strain of E. coli, AHL is not produced without IPTG induction, but luxR expression is controlled by the constitutive ptet promoter. We hypothesize that since luxR was present in these cells without AHL, the backward transcription from luxPR hindered the function of the terminators that separated the luxR gene from the GFP gene. Thus GFP may have been expressed because the ptet promoter, which is constitutive, was able to affect its expression. | ||
| + | |||
| + | In order to bypass this issue, we are working to insert two more terminators between the luxR gene and the luxPR promoter, in hopes that this will stop the backward transcription from having an effect on GFP expression, allowing us to work towards measuring the time delay created by our construct. | ||
| + | |||
| + | |||
| + | <h1>Characterization of Promoter. Improvement by BIT-China 2019</h1> | ||
| + | To see the leakage of the promoter, we performed a 3 hour fluorescence characterization of bba_k1499500 part. The experimental results showed that the promoter leakage degree of pLUX increased with the culture time. | ||
| + | |||
| + | The results are shown below:<br> | ||
| + | [[File:Yfc 111111.png|center|thumb|400px|<b>Fig.Fluorescence fermentation results of BBa_K1499500.</b>]] | ||
| + | |||
| + | To make it more suitable for our project and its wider application, we have characterized it and improved it.<br> | ||
| + | Please see BBa_K2972008 for details about the Improvement.<br> | ||
<!-- --> | <!-- --> | ||
| Line 18: | Line 43: | ||
<partinfo>BBa_K1499500 SequenceAndFeatures</partinfo> | <partinfo>BBa_K1499500 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | == References== | ||
| + | [http://2010.igem.org/Team:Cambridge/ProjectBioluminescence/Luciferase/WikiGeneticsLuxCDABE] | ||
| + | [https://parts.igem.org/wiki/index.php?title=Part:BBa_R0062] | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 03:37, 22 October 2019
quorum sensing machinery that activates GFP expression
This part is the combination of two parts, BBa_I13202 and BBa_T9002. The first part (BBa_I13202) produces a luxI enzyme which produces an AHL (3OC6HSL). This part is under the control of a lac repressible promoter. The second part (BBa_T9002) produces luxR, which combines with the AHL to activate the luxPR promoter that controls expression of GFP (see image below [1]).
Our goal in building this construct is to create a time delay. Since the production of AHL is under the control of a lac-repressible promoter, the cascade should not begin until the promoter is induced. Additionally, once the promoter is activated, time is required for the AHL and luxR molecules to build up and interact with one another. Only after these molecules are present in large enough quantities to interact and bind to the luxPR promoter will GFP be expressed. Thus by replacing GFP with any gene of interest, this construct can be used to create a time delay for the onset of gene expression. For more information on the specifics of the time delay, see the "experience" page.
Usage and Biology
After ligating this construct together and into the BioBrick backbone, we transformed it into the NEB 5-alpha strain of E. coli. The colonies that fluoresced were most likely to have been successfully transformed, and thus these were sent off for sequencing. The sequencing data showed that our construct was correct (see below), so we were able to submit for BioBricking.
Although our sequencing data was good, we realized that we had not induced the lac-repressible promoter, and thus the colonies should not have been fluorescing yet. After doing more research, we realized that the strain of E. coli we were using was not lactose deficient, thus the quorum sensing cascade was being activated by the natural presence of lactose. We ordered a lacI^q strain of E. coli, which are lac deficient, so that we could have complete control of GFP expression. However, when we performed the transformation of the construct into this strain, all of the colonies still fluoresced.
After doing further research, we discovered that the luxPR promoter (BBa_R0062) [2], will induce backward transcription if the luxR protein is present but the AHL molecule is not. In the lac deficient strain of E. coli, AHL is not produced without IPTG induction, but luxR expression is controlled by the constitutive ptet promoter. We hypothesize that since luxR was present in these cells without AHL, the backward transcription from luxPR hindered the function of the terminators that separated the luxR gene from the GFP gene. Thus GFP may have been expressed because the ptet promoter, which is constitutive, was able to affect its expression.
In order to bypass this issue, we are working to insert two more terminators between the luxR gene and the luxPR promoter, in hopes that this will stop the backward transcription from having an effect on GFP expression, allowing us to work towards measuring the time delay created by our construct.
Characterization of Promoter. Improvement by BIT-China 2019
To see the leakage of the promoter, we performed a 3 hour fluorescence characterization of bba_k1499500 part. The experimental results showed that the promoter leakage degree of pLUX increased with the culture time.
The results are shown below:
To make it more suitable for our project and its wider application, we have characterized it and improved it.
Please see BBa_K2972008 for details about the Improvement.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1815
Illegal BsaI.rc site found at 2543
References
[http://2010.igem.org/Team:Cambridge/ProjectBioluminescence/Luciferase/WikiGeneticsLuxCDABE] [1]