Difference between revisions of "Part:BBa K3304105"
PlatonMega (Talk | contribs) |
|||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3304105 short</partinfo> | <partinfo>BBa_K3304105 short</partinfo> | ||
| − | This is the Input Gate for our Chemical Reaction Network. The oligo input is DTG (part: BBa_K3304118 ) wich initiates the strand displacement cascades. Other auxiliary strands for this gate are BTD (part: BBa_K3304108 ) and NTB (part: BBa_K3304109 ) who conclude the reactions and facilitate their trajectory to the desired direction. The output of this gate is TBB (part: BBa_K3304110 ) which constitutes the input for the downstream Join Gate (part: BBa_K3304106 ). The part comes with two PvuII cut sites and three Nt.BstNBI nicking sites that facilitate the extraction of the gate from plasmid DNA and prepare the double-stranded block for strand displacement reactions. | + | This is the Input Gate for our Chemical Reaction Network. The oligo input is DTG (part: BBa_K3304118 ) wich initiates the strand displacement cascades. Other auxiliary strands for this gate are BTD (part: BBa_K3304108 ) and NTB (part: BBa_K3304109 ) who conclude the reactions and facilitate their trajectory to the desired direction. The output of this gate is TBB (part: BBa_K3304110 ) which constitutes the input for the downstream Join Gate (part: BBa_K3304106 ). The part comes with two PvuII cut sites and three Nt.BstNBI nicking sites that facilitate the extraction of the gate from plasmid DNA and prepare the double-stranded block for strand displacement reactions. The ELK-1s binding site is incorporated at the TDD domain allowing for the ELK-1 TF to bind to the sequence, thus inhibiting the initiation of strand displacement reactions, reducing the concentration of available Input Gates and subsequently the amount input TBB, resulting in proportional circuit behavior. |
https://2019.igem.org/wiki/images/c/ca/T--Thessaloniki--animation-step1.gif | https://2019.igem.org/wiki/images/c/ca/T--Thessaloniki--animation-step1.gif | ||
Revision as of 03:19, 22 October 2019
Input Gate with ELK1 sequence
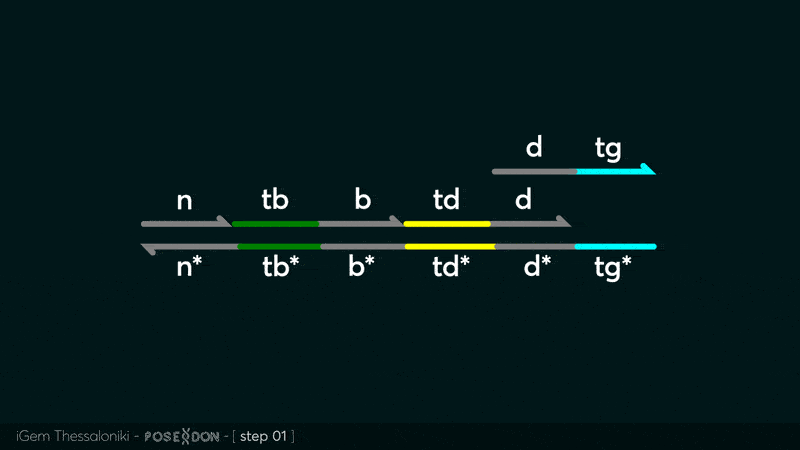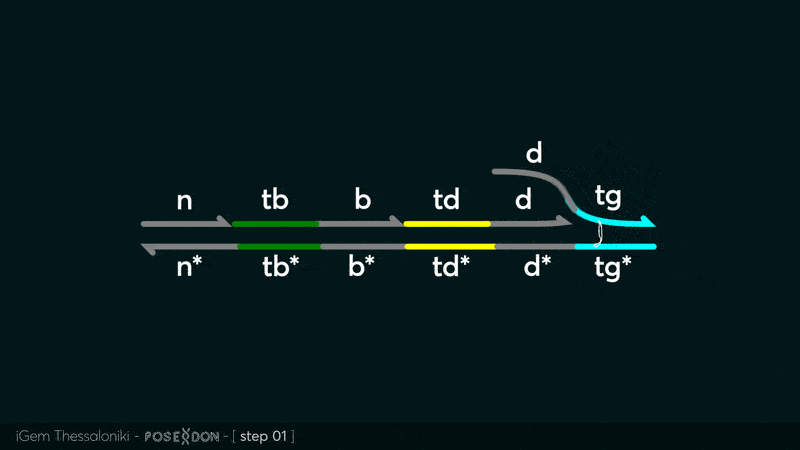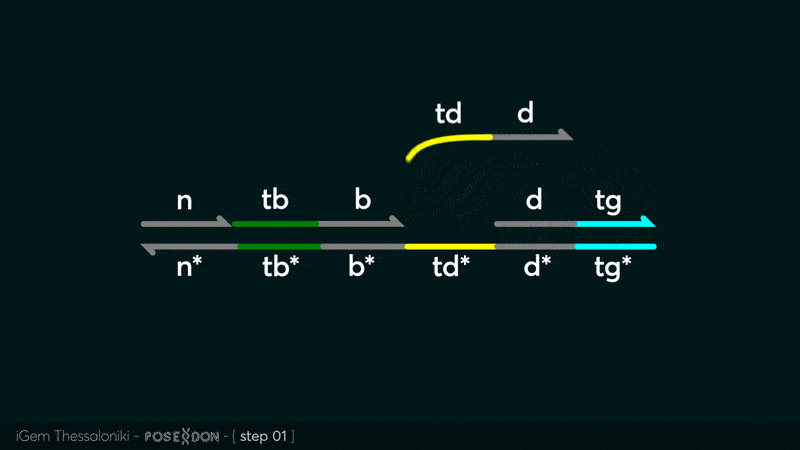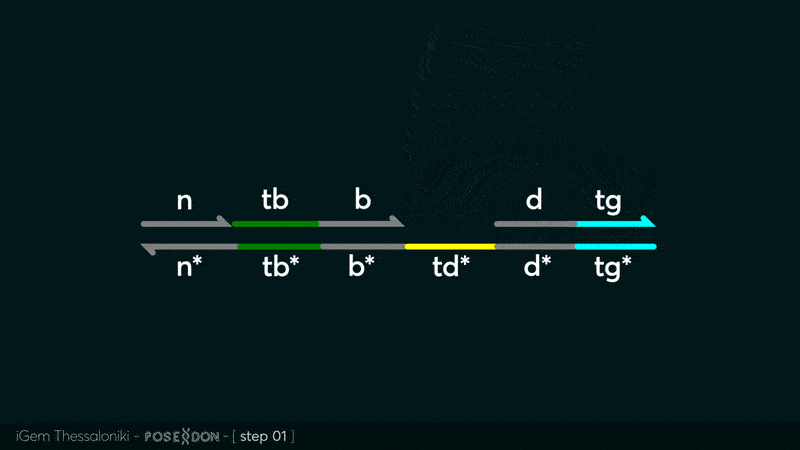
This is the Input Gate for our Chemical Reaction Network. The oligo input is DTG (part: BBa_K3304118 ) wich initiates the strand displacement cascades. Other auxiliary strands for this gate are BTD (part: BBa_K3304108 ) and NTB (part: BBa_K3304109 ) who conclude the reactions and facilitate their trajectory to the desired direction. The output of this gate is TBB (part: BBa_K3304110 ) which constitutes the input for the downstream Join Gate (part: BBa_K3304106 ). The part comes with two PvuII cut sites and three Nt.BstNBI nicking sites that facilitate the extraction of the gate from plasmid DNA and prepare the double-stranded block for strand displacement reactions. The ELK-1s binding site is incorporated at the TDD domain allowing for the ELK-1 TF to bind to the sequence, thus inhibiting the initiation of strand displacement reactions, reducing the concentration of available Input Gates and subsequently the amount input TBB, resulting in proportional circuit behavior.
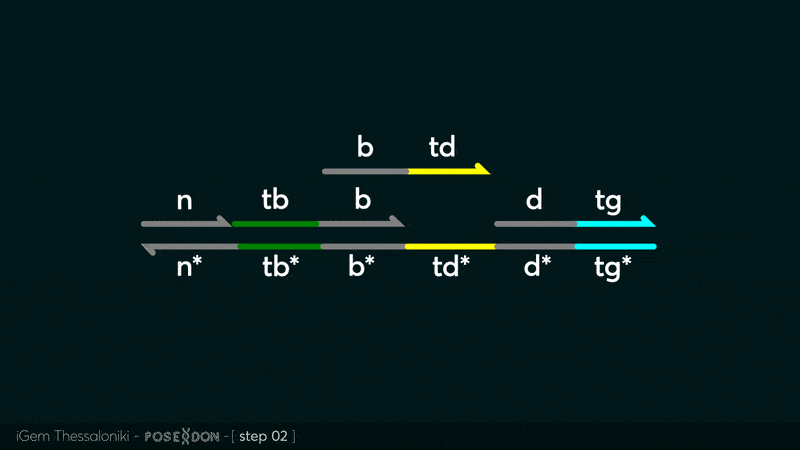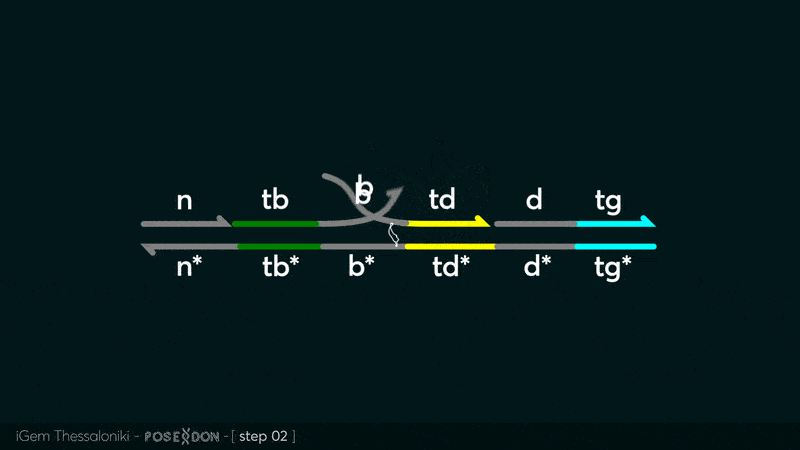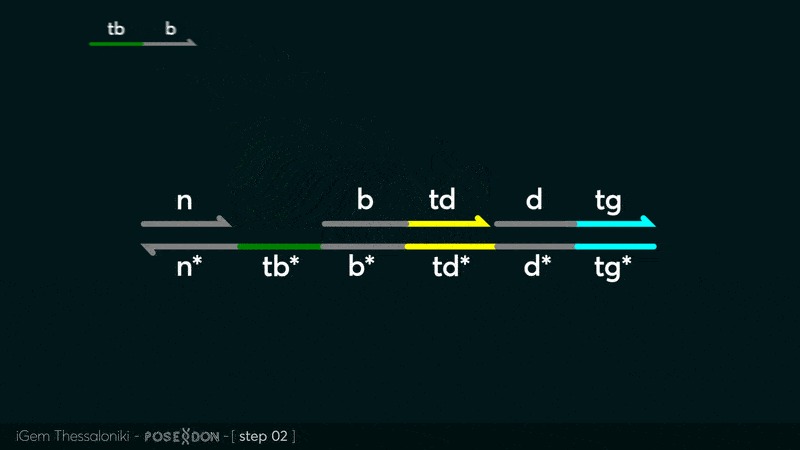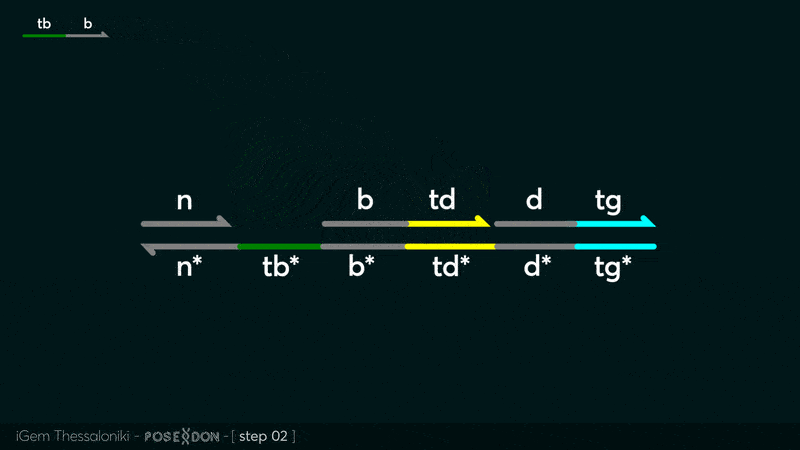

 https://2019.igem.org/File:T--Thessaloniki--results-f12.png
Sequence and Features
https://2019.igem.org/File:T--Thessaloniki--results-f12.png
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
