Difference between revisions of "Part:BBa K3036008"
| (One intermediate revision by one other user not shown) | |||
| Line 50: | Line 50: | ||
6.Three replicas are tested in each group. <br> | 6.Three replicas are tested in each group. <br> | ||
| − | <font size="4"><b> | + | <b><font size="3">Reference</font></b> |
| − | <font size="3">We imporved <html><a href="https://parts.igem.org/Part:BBa_K185048" target="_blank">BBa_K185048</a> </font> | + | |
| + | [1] Butz M., Neuenschwander M., Kast P., Hilvert D.. An N-Terminal Protein Degradation Tag Enables Robust Selection of Highly Active Enzymes [J]. Biochemistry, 2011, 50(40):8594-8602. | ||
| + | |||
| + | |||
| + | <font size="4"><b>Improved from BBa_K185048 </b></font><br> | ||
| + | <font size="3">We imporved <html><a href="https://parts.igem.org/Part:BBa_K185048" target="_blank">BBa_K185048</a> </font> | ||
Latest revision as of 02:09, 22 October 2019
RelB with degradation-promoting tag RepA
RelB is the inhibitor of toxin RelE. The two proteins can be made use of in a toxin-antitoxin system, where RelE gets constantly expressed and RelB is expressed only under certain conditions, validating a kill switch that is triggered when its host escapes to the outer environment.
Biology and Usage
We improve antitoxin RelB (BBa_K185048) by promoting its degradation rate so that the kill switch where it is involved functions at a more rapid pace, preventing contamination with higher efficiency. RelB is the antitoxin for toxin RelE (BBa_K185047). RelE inhibits protein synthesis of bacteria by cleaving mRNAs at A site of ribosomes, and RelB functions by binding to RelE and thereby relieves its toxicity. Such a toxin-antitoxin system can be readily used in a biosafety system, where RelE is constantly expressed while RelB gets expressed only under certain permissive conditions. Consequently, when the bacteria escapes from the conditions, RelB stops expressing, causing excess of RelE and thus the death of escaped microbes. Therefore, the degradation rate of RelB is decisive to the performance of this system.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Properties
We improve the function of RelB by increasing its degradation rate so that once the expression stops, RelB degrades at a more rapid pace and leads to death of bacteria in a shorter period of time, which is a critical trait when assessing the validity of a biosafety system. To promote the degradation of RelB, we add a degradation-promoting tag RepA to RelB. RepA is a 16-amino-acid long peptide that conducts degradation of protein using native machinery in E. coli.
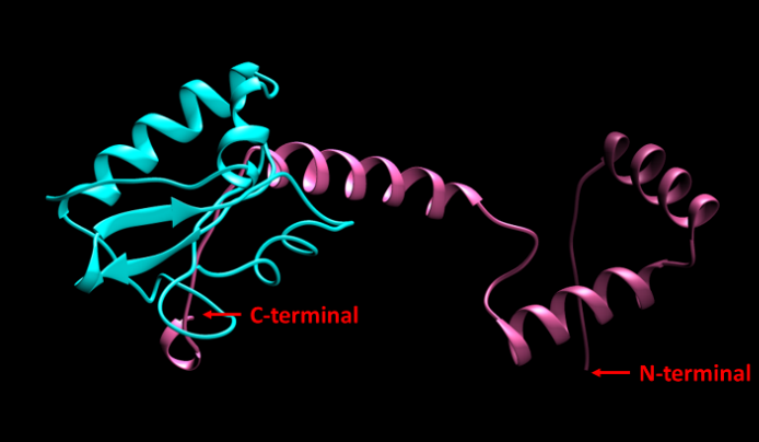
In our improvement, RepA is placed at N-terminal of RelB. This strategy has two advantages: first, as is shown in Figure 1, the interaction with RelE is performed by the C-terminal domain of RelB, as a result, addition of degradation tag at N-terminal does not interfere with its function; second, adding the tag at N-terminal of RelB prevents the protein from losing the peptide through nonsense mutation.
Results
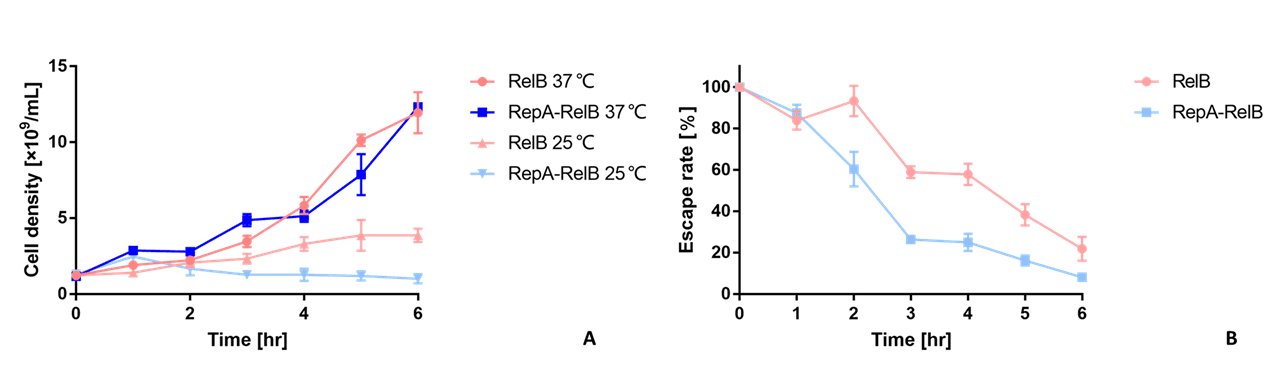
We compare the function of RelB before and after improvement using the toxin-antitoxin system. As is shown in Figure 2, under same nonpermissive conditions, the bacteria containing the kill switch with improved RelB die at a notably higher rate than those with unimproved RelB, verifying the effect of the improvement. Otherwise, the OD600 shows no notable difference, indicating RelE kills bacteria without lysing the cell.
Experimental Approaches
1.Transform kill switch systems with RelB or RepA-RelB into E. coli DH5alpha competent cells respectively.
2.Culture both strains with LB medium at 37℃ overnight.
3.Equally divide each culture into two flasks. Put one of them at 37℃ and the other at 27℃.
4.Extract 5μl samples of each culture system every 6 hours. Diluted the samples 10^7 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately.
5.Count the number of colonies in 5 cm^2 per plate after cultured for 24 hours at 37℃.
6.Three replicas are tested in each group.
Reference
[1] Butz M., Neuenschwander M., Kast P., Hilvert D.. An N-Terminal Protein Degradation Tag Enables Robust Selection of Highly Active Enzymes [J]. Biochemistry, 2011, 50(40):8594-8602.
Improved from BBa_K185048
We imporved BBa_K185048