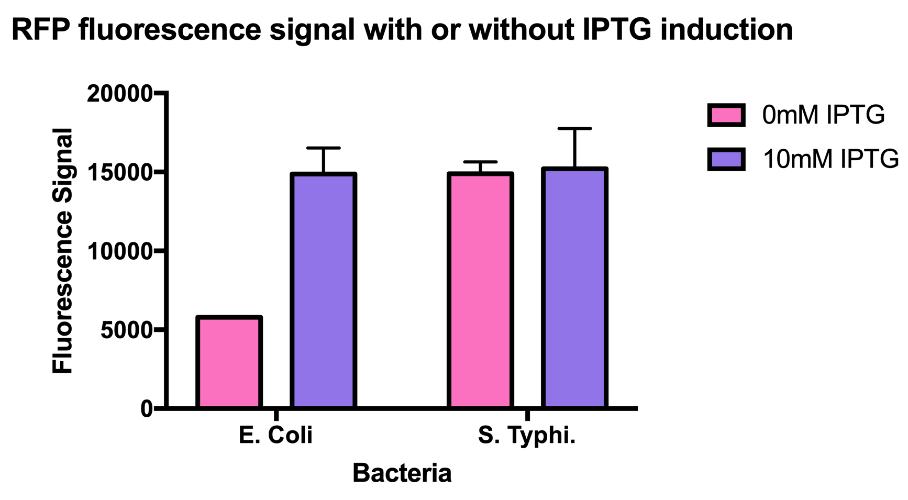Difference between revisions of "Part:BBa R0010"
(r) |
|||
| Line 34: | Line 34: | ||
[http://2016.igem.org/Team:Tec-Monterrey Team Tec-Monterrey 2016] tested the Promoter BBa_R0010 in <i>C. Violaceum</i>. We were unable to confirm whether or not <i>C. Violaceum</i> had the lac operon, BLAST analysis of its genome suggested that it did not have this regulating system; our characterization demonstrates that the lac promoter can be used as a constitutive promoter in this chassis. There was no significant difference in the Fluorescence of the group of <i>C. Violaceum</i> with IPTG or without it, but in <i>E. Coli</i> it showed normal induction. Therefore, we can confirm that this promoter is constitutive in <i>C. Violaceum</i>. https://static.igem.org/mediawiki/parts/4/4b/T--Tec-Monterrey--violaceumgrafiicauno.jpg https://static.igem.org/mediawiki/parts/7/76/T--Tec-Monterrey--violaceumgrafiicados.jpg | [http://2016.igem.org/Team:Tec-Monterrey Team Tec-Monterrey 2016] tested the Promoter BBa_R0010 in <i>C. Violaceum</i>. We were unable to confirm whether or not <i>C. Violaceum</i> had the lac operon, BLAST analysis of its genome suggested that it did not have this regulating system; our characterization demonstrates that the lac promoter can be used as a constitutive promoter in this chassis. There was no significant difference in the Fluorescence of the group of <i>C. Violaceum</i> with IPTG or without it, but in <i>E. Coli</i> it showed normal induction. Therefore, we can confirm that this promoter is constitutive in <i>C. Violaceum</i>. https://static.igem.org/mediawiki/parts/4/4b/T--Tec-Monterrey--violaceumgrafiicauno.jpg https://static.igem.org/mediawiki/parts/7/76/T--Tec-Monterrey--violaceumgrafiicados.jpg | ||
| + | |||
| + | ==Characterization of expression in different sugars mixtures== | ||
| + | |||
| + | <html> | ||
| + | <p> | ||
| + | See more results on <a style="color:#3BB9FF;" href=« https://2019.igem.org/Team:Nantes/Results">our wiki page</a>. | ||
| + | </p> | ||
| + | <br> | ||
| + | |||
| + | |||
| + | <div> | ||
| + | <br> | ||
| + | <h1 style="text-align:center;">I - Testing of the functionality of our designed parts</h1> | ||
| + | <br> | ||
| + | <p>After transforming K12 MG1655 <span class="italic">E.coli</span> with our 6 different constructs, we measured by spectrofluorometry the fluorescence of the transformed bacteria cultivated in M9 medium in the presence of saturating concentration | ||
| + | (0.2%) of the associated sugar . The fluorescence studied in the cases below has been normalized with the Optical Density (OD).</p> | ||
| + | <br> | ||
| + | <p>Our spectrofluorometric measurements were carried out on the transformed bacteria cultivated in a M9 medium in 2 different conditions : with 0.2% of the specific sugar, and without sugar.</p> | ||
| + | <div> | ||
| + | <br> | ||
| + | |||
| + | <div> | ||
| + | <h4 style="text-decoration: underline;"> ● pSRL-RFP <span class="italic">E.coli</span> :</h4> | ||
| + | <br> | ||
| + | <img class="imagegraph" src="https://2019.igem.org/wiki/images/6/6f/T--Nantes--graph3.png" alt=""> | ||
| + | <p><span class="bold">Figure 3:</span> RFP Fluorescence normalized with OD for <span class="italic">E.coli</span> K12 MG1655 transformed with pSRL in the presence (red) or absence (grey) of sorbitol at 0,2%. Experiments were conducted during | ||
| + | 17 hours.</p> | ||
| + | <p> | ||
| + | The activity of pSRL is relative to the expression of RFP fluorescence. We do not observe a significant difference of RFP fluorescence between both conditions, with or without sorbitol. Either (i) there is a RFP fluorescence in both conditions meaning | ||
| + | that the promoter pSRL is active even in the absence of sugar, or (ii) we are in presence of an experimental error. | ||
| + | </p> | ||
| + | </div> | ||
| + | <br> | ||
| + | <div> | ||
| + | <h2 style="text-align:center;">Summary</h2> | ||
| + | <p> | ||
| + | Unfortunately, we were not able to have pSRL construct to work properly. </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <br> | ||
| + | <!--Partie 2--> | ||
| + | <br> | ||
| + | <div> | ||
| + | <h1 style="text-align:center;">II - The link between sugar concentration and promoter activity</h1> | ||
| + | <br> | ||
| + | <p>For these tests we measured the fluorescence in K12 MG1655 <span class="italic">E.coli</span> transformed with our pSRL-RFP. The bacteria were cultivated in M9 medium with various different concentrations of the associated sugar. The fluorescence studied in the graphs below has been normalised with the Optical Density (OD). | ||
| + | |||
| + | </p> | ||
| + | <br> | ||
| + | <br> | ||
| + | <div> | ||
| + | <h4 style="text-decoration: underline;"> ● pSRL-RFP <span class="italic">E.coli</span> :</h4> | ||
| + | <img class="imagegraph" style="margin-left:10%;width:80%; height:80%;" src="https://2019.igem.org/wiki/images/d/dd/T--Nantes--graph11.png" alt=""> | ||
| + | <p><span class="bold">Figure 11:</span> RFP Fluorescence normalized with OD for <span class="italic">E.coli</span> K12 MG1655 transformed with pSRL in selected concentrations of Sorbitol | ||
| + | </p> | ||
| + | <p> | ||
| + | The activity of pSRL is relative to the expression of RFP fluorescence. This figure shows no significant difference of RFP expression between all the different conditions This is coherent with our previous observations whereby we observed that pSRL activity | ||
| + | was equivalent in absence and in presence of 0.2% sorbitol (see figure 3). The pSRL does not seem to be affected by the sugar in the medium, therefore we assume a dysfunctionality of this promoter in our constructs | ||
| + | </p> | ||
| + | </div> | ||
| + | <br> | ||
| + | |||
| + | <div> | ||
| + | <h2 style="text-align:center;">Summary</h2> | ||
| + | <p> | ||
| + | Unfortunately, we were not able to have pSRL construct to work properly. <br> | ||
| + | </p> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
==Characterization for usage in genetically recoded organisms and expanded genetic code== | ==Characterization for usage in genetically recoded organisms and expanded genetic code== | ||
Revision as of 01:15, 22 October 2019
promoter (lacI regulated)
This part is an inverting regulator sensitive to LacI and CAP.
It contains two protein binding sites. The first binds the CAP protein, which is generally present in E.coli and is asocciated with cell health and availability of glucose. The second binds LacI protein.
- In the absence of LacI protein and CAP protein, this part promotes transcription.
- In the presence of LacI protein and CAP protein, this part inhibits transcription.
- LacI can be inhibited by [http://openwetware.org/wiki/IPTG IPTG].
- LacI is coded by BBa_C0010
Intrinsic noise value: 0.0707 (compare with R0011: 0.0040; R0051: 0.0869). See [http://2015.igem.org/Team:William_and_Mary William_and_Mary iGEM 2015]
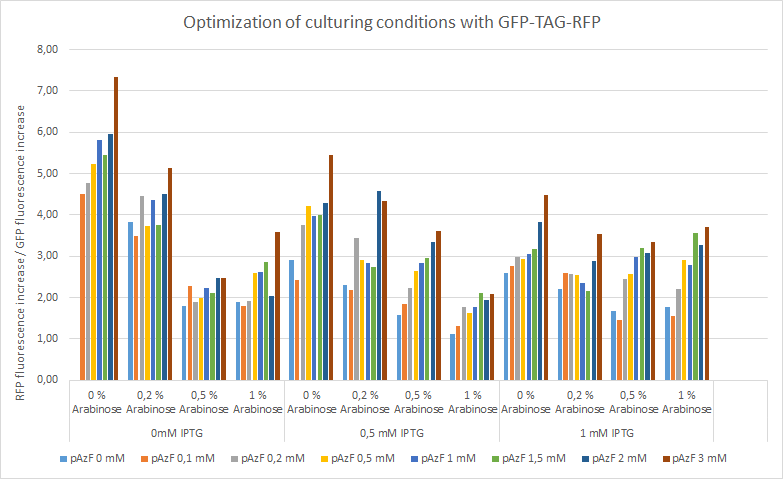
>Internal Priming Screening Characterization of BBa_R0010: Has 3 possible internal priming site between this BioBrick part and the VR primer.
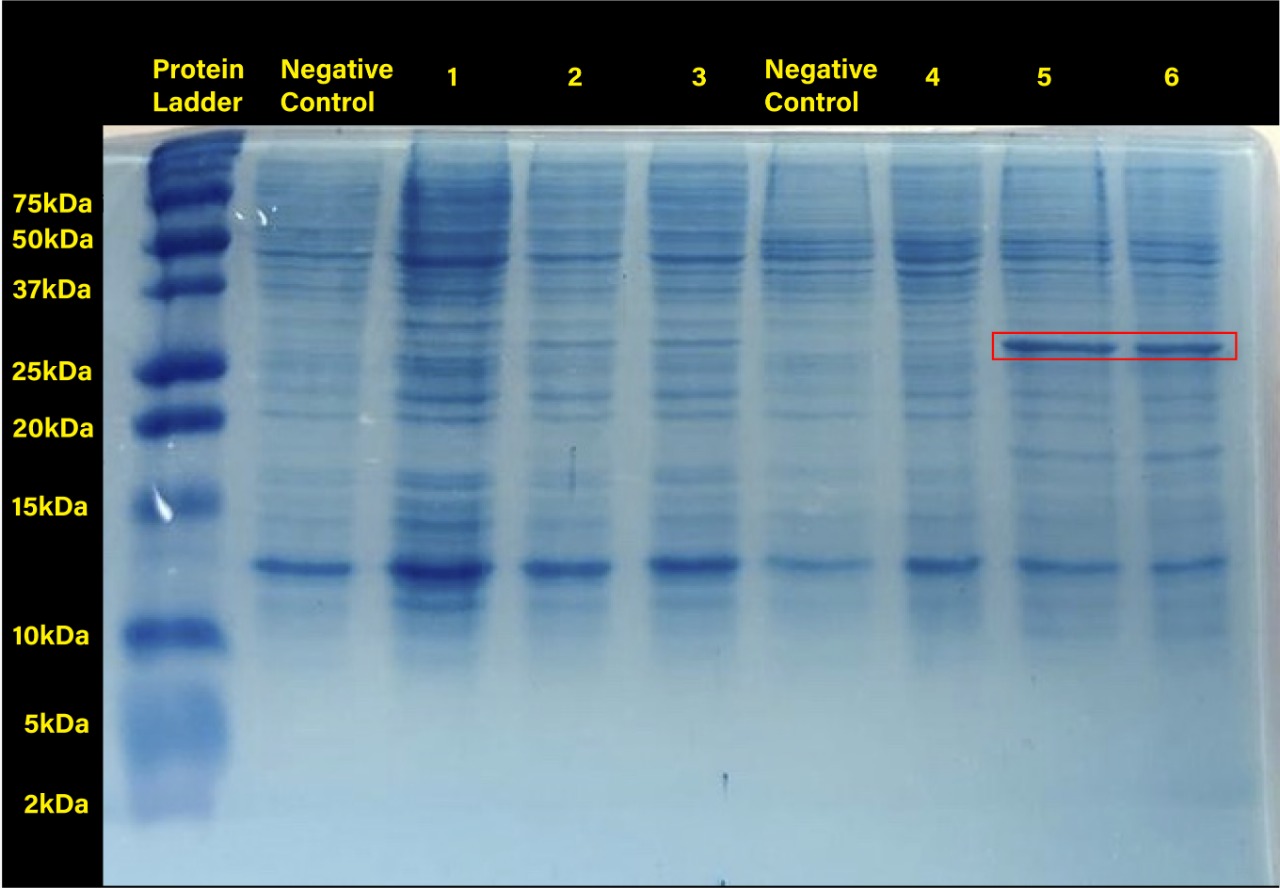
The 2018 Hawaii iGEM team evaluated the 40 most frequently used BioBricks and ran them through an internal priming screening process that we developed using the BLAST program tool. Out of the 40 BioBricks we evaluated, 10 of them showed possible internal priming of either the VF2 or VR primers and sometime even both. The data set has a range of sequence lengths from as small as 12 bases to as large as 1,210 bases. We experienced the issue of possible internal priming during the sequence verification process of our own BBa_K2574001 BioBrick and in the cloning process to express the part as a fusion protein. BBa_K2574001 is a composite part containing a VLP forming Gag protein sequence attached to a frequently used RFP part (BBa_E1010). We conducted a PCR amplification of the Gag-RFP insert using the VF2 and VR primers on the ligation product (pSB1C3 ligated to the Gag + RFP). This amplicon would serve as template for another PCR where we would add the NcoI and BamHI restriction enzyme sites through new primers for ligation into pET14b and subsequent induced expression. Despite gel confirming a rather large, approximately 2.1 kb insert band, our sequencing results with the VR primer and BamHI RFP reverse primer gave mixed results. Both should have displayed the end of the RFP, but the VR primer revealed the end of the Gag. Analysis of the VR primer on the Gag-RFP sequence revealed several sites where the VR primer could have annealed with ~9 - 12 bp of complementarity. Internal priming of forward and reverse primers can be detrimental to an iGEM project because you can never be sure if the desired construct was correctly inserted into the BioBrick plasmid without a successful sequence verification.
For the BioBrick part BBa_R0010, the first location of the internal priming site is on the 121-113 base number of the BioBrick and on the 12-20 base number of the VR primer. The second location of the internal priming site is on the 11-17 base number of the BioBrick and on the 4-10 base number of the VR primer. The third location of the internal priming site is on the 84-90 base number of the BioBrick and on the 14-20 base number of the VR primer.
Usage and Biology
This is a direct copy of bases 0365739 through 0365540 of the E. coli K-12 MG1655 genome, Genbank NC_000913 in reverse complement form. It is the natural promoter for the LacZYA operon. It includes the tail end of the LacI gene coding region, but no promoter region for that partial gene.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage in Chromobacterium Violaceum
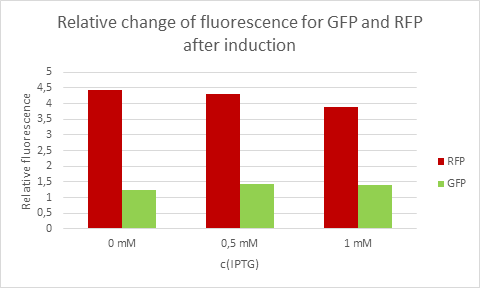
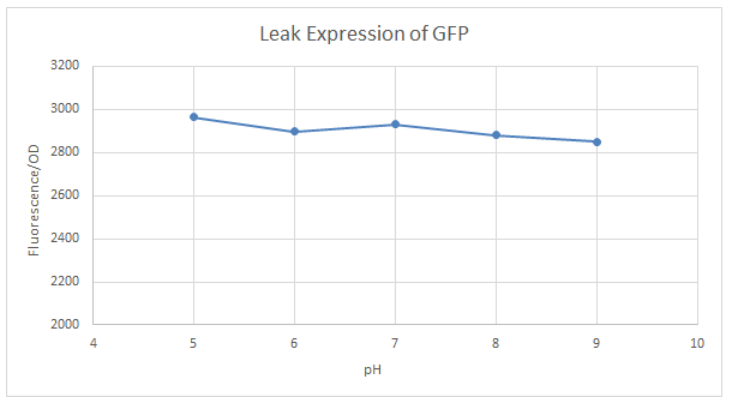
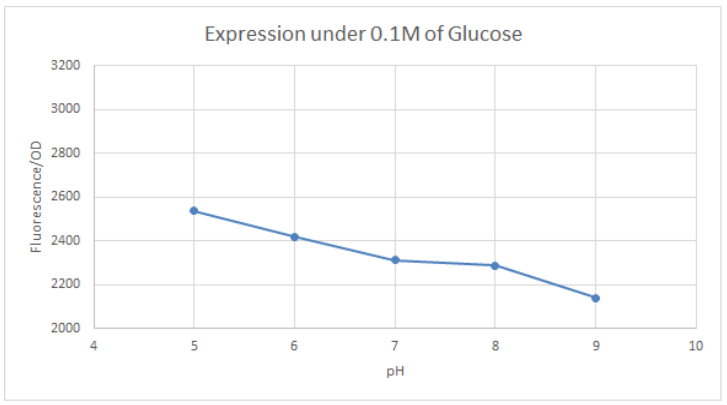
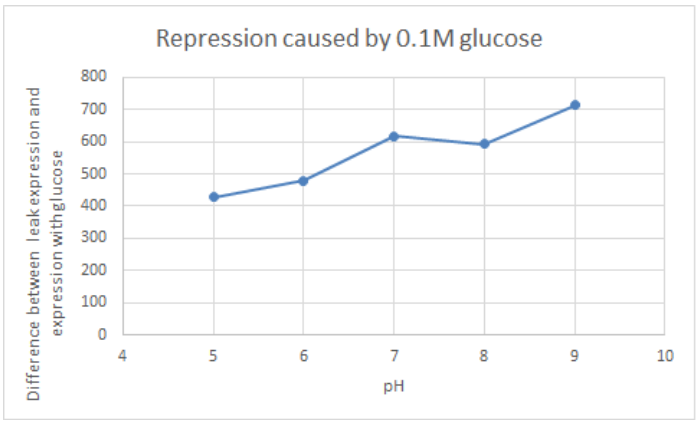
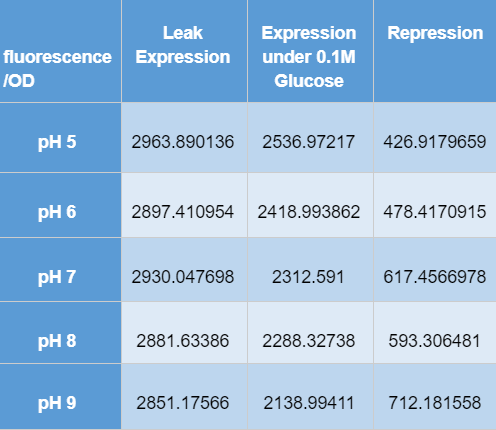
[http://2016.igem.org/Team:Tec-Monterrey Team Tec-Monterrey 2016] tested the Promoter BBa_R0010 in C. Violaceum. We were unable to confirm whether or not C. Violaceum had the lac operon, BLAST analysis of its genome suggested that it did not have this regulating system; our characterization demonstrates that the lac promoter can be used as a constitutive promoter in this chassis. There was no significant difference in the Fluorescence of the group of C. Violaceum with IPTG or without it, but in E. Coli it showed normal induction. Therefore, we can confirm that this promoter is constitutive in C. Violaceum. 

Characterization of expression in different sugars mixtures
See more results on our wiki page.
I - Testing of the functionality of our designed parts
After transforming K12 MG1655 E.coli with our 6 different constructs, we measured by spectrofluorometry the fluorescence of the transformed bacteria cultivated in M9 medium in the presence of saturating concentration (0.2%) of the associated sugar . The fluorescence studied in the cases below has been normalized with the Optical Density (OD).
Our spectrofluorometric measurements were carried out on the transformed bacteria cultivated in a M9 medium in 2 different conditions : with 0.2% of the specific sugar, and without sugar.
● pSRL-RFP E.coli :

Figure 3: RFP Fluorescence normalized with OD for E.coli K12 MG1655 transformed with pSRL in the presence (red) or absence (grey) of sorbitol at 0,2%. Experiments were conducted during 17 hours.
The activity of pSRL is relative to the expression of RFP fluorescence. We do not observe a significant difference of RFP fluorescence between both conditions, with or without sorbitol. Either (i) there is a RFP fluorescence in both conditions meaning that the promoter pSRL is active even in the absence of sugar, or (ii) we are in presence of an experimental error.
Summary
Unfortunately, we were not able to have pSRL construct to work properly.
II - The link between sugar concentration and promoter activity
For these tests we measured the fluorescence in K12 MG1655 E.coli transformed with our pSRL-RFP. The bacteria were cultivated in M9 medium with various different concentrations of the associated sugar. The fluorescence studied in the graphs below has been normalised with the Optical Density (OD).
● pSRL-RFP E.coli :

Figure 11: RFP Fluorescence normalized with OD for E.coli K12 MG1655 transformed with pSRL in selected concentrations of Sorbitol
The activity of pSRL is relative to the expression of RFP fluorescence. This figure shows no significant difference of RFP expression between all the different conditions This is coherent with our previous observations whereby we observed that pSRL activity was equivalent in absence and in presence of 0.2% sorbitol (see figure 3). The pSRL does not seem to be affected by the sugar in the medium, therefore we assume a dysfunctionality of this promoter in our constructs
Summary
Unfortunately, we were not able to have pSRL construct to work properly.