Difference between revisions of "Part:BBa K3190601"
Hitesh Gelli (Talk | contribs) |
|||
| (2 intermediate revisions by 2 users not shown) | |||
| Line 10: | Line 10: | ||
We wanted to improve the BI-1 coding sequence (<partinfo>BBa_K2365518</partinfo>), which was submitted by the iGEM team NAU_CHINA 2017. They used it as part of their kill switch system and also submitted a composite part consisting of the TEF1 constitutive promoter, BI-1, and the yeast CYC1 terminator. | We wanted to improve the BI-1 coding sequence (<partinfo>BBa_K2365518</partinfo>), which was submitted by the iGEM team NAU_CHINA 2017. They used it as part of their kill switch system and also submitted a composite part consisting of the TEF1 constitutive promoter, BI-1, and the yeast CYC1 terminator. | ||
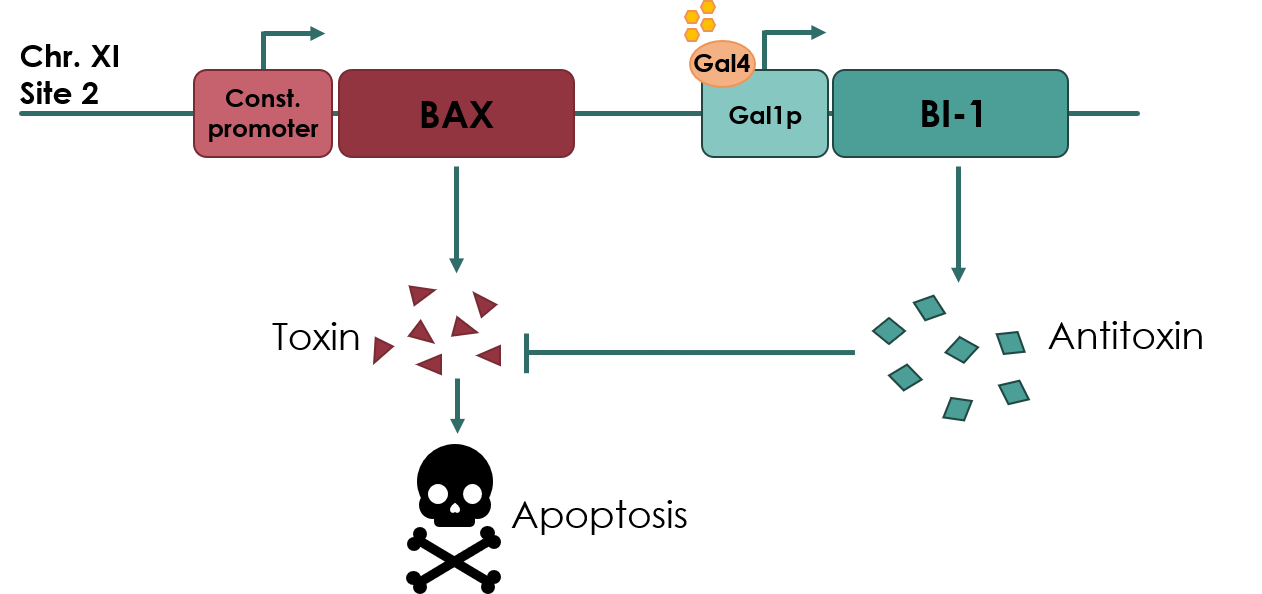
| − | The kill switch proposed by team NAU China | + | The kill switch proposed by team NAU China would work by co-expressing the BAX gene (<partinfo>BBa_K2365048</partinfo>), which encodes the pro-apoptotic BAX protein, under a constitutive promoter, together with BI-1 under the galactose inducible GAL1 promoter. In theory, as long as galactose is present in the media, BI-1 will be expressed, inhibiting the BAX protein. Should the yeast cell escape the media or matrix, BAX will be expressed, causing apoptosis of the cell. |
[[File:UCopenhagen Killswitch.jpeg|600px]] | [[File:UCopenhagen Killswitch.jpeg|600px]] | ||
| Line 16: | Line 16: | ||
<small><b>Figure 1: Overview of the kill switch </b></small> | <small><b>Figure 1: Overview of the kill switch </b></small> | ||
| − | + | We improved the existing BI-1 sequence by expressing it under the control of the inducible pGAL1 promoter. This improvement will allow us to control the expression of BI-1, as the GAL1 promoter allows transcription of BI-1 only in the presence of galactose. | |
| Line 22: | Line 22: | ||
In order to show that BI-1 is able to neutralize the lethal impact of BAX, we performed a rescue assay in which both BAX and BI-1 were placed under the control of the GAL1 promoter. For this, we constructed two strains; OV19, which had pGAL1-BAX integrated into the genome and pGAL1-BI-1 on a high-copy number episomal plasmid, and OV20, which had pGAL1-BAX integrated into the genome and carried an empty vector. | In order to show that BI-1 is able to neutralize the lethal impact of BAX, we performed a rescue assay in which both BAX and BI-1 were placed under the control of the GAL1 promoter. For this, we constructed two strains; OV19, which had pGAL1-BAX integrated into the genome and pGAL1-BI-1 on a high-copy number episomal plasmid, and OV20, which had pGAL1-BAX integrated into the genome and carried an empty vector. | ||
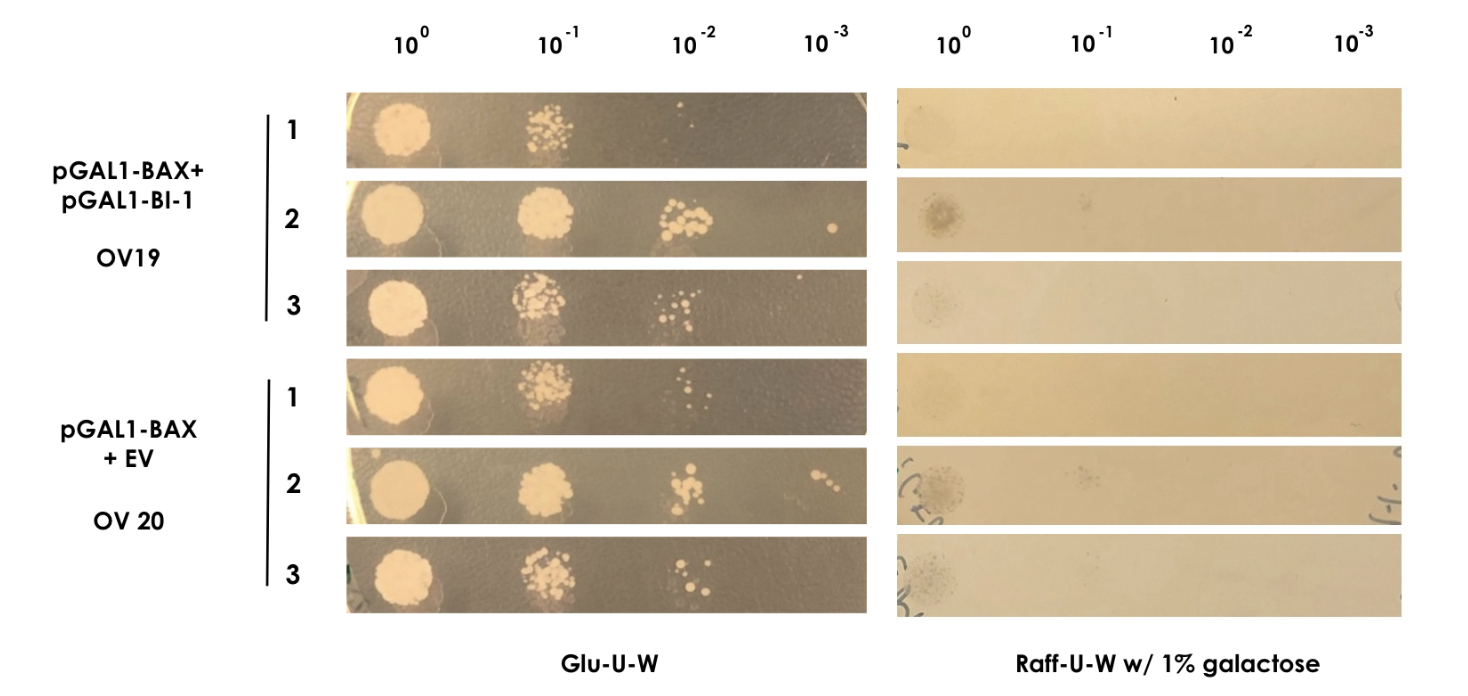
| − | For the assay colonies were picked and diluted in sterile water. Using the comparative galactose induction assay as a template, the ODs at 600 nm were equalized to 0.035 and different dilutions of the colonies (10 | + | For the assay colonies were picked and diluted in sterile water. Using the comparative galactose induction assay as a template, the ODs at 600 nm were equalized to 0.035 and different dilutions of the colonies (10<sup>-0</sup> to 10<sup>-3</sup>) were spotted on plates with glu-U-W agar and raff-U-W agar with 1% galactose in volumes of 10 µl. A yeast strain containing only pGAL1-BAX and the empty pWUS plasmid served as control (OV20) and was spotted along with the sample (Figure 2). |
| − | [[File:ovulaid35.png| | + | [[File:ovulaid35.png|800px]] |
<small><b>Figure 2: BAX rescue assay</b> | In this assay, the strains OV19 and OV20 were grown on both glu-U-W agar (left) and raff-U-W (1% galactose) agar in different dilutions of OD600nm (10-0 to 10-3). </small> | <small><b>Figure 2: BAX rescue assay</b> | In this assay, the strains OV19 and OV20 were grown on both glu-U-W agar (left) and raff-U-W (1% galactose) agar in different dilutions of OD600nm (10-0 to 10-3). </small> | ||
Latest revision as of 00:27, 22 October 2019
pGAL1-BI-I: BI-I CDS under inducible promoter
Coding sequence of BAX Inhibitor-I (BI-1) anti-toxin under control of the galactose inducible GAL1 promoter (BBa_K3190050). This biobrick was designed for use in a yeast kill switch system, as described by NAU_CHINA 2017 (Figure 1).
Usage and Biology
Improvement of kill switch system
We wanted to improve the BI-1 coding sequence (BBa_K2365518), which was submitted by the iGEM team NAU_CHINA 2017. They used it as part of their kill switch system and also submitted a composite part consisting of the TEF1 constitutive promoter, BI-1, and the yeast CYC1 terminator.
The kill switch proposed by team NAU China would work by co-expressing the BAX gene (BBa_K2365048), which encodes the pro-apoptotic BAX protein, under a constitutive promoter, together with BI-1 under the galactose inducible GAL1 promoter. In theory, as long as galactose is present in the media, BI-1 will be expressed, inhibiting the BAX protein. Should the yeast cell escape the media or matrix, BAX will be expressed, causing apoptosis of the cell.
Figure 1: Overview of the kill switch
We improved the existing BI-1 sequence by expressing it under the control of the inducible pGAL1 promoter. This improvement will allow us to control the expression of BI-1, as the GAL1 promoter allows transcription of BI-1 only in the presence of galactose.
BAX rescue assay
In order to show that BI-1 is able to neutralize the lethal impact of BAX, we performed a rescue assay in which both BAX and BI-1 were placed under the control of the GAL1 promoter. For this, we constructed two strains; OV19, which had pGAL1-BAX integrated into the genome and pGAL1-BI-1 on a high-copy number episomal plasmid, and OV20, which had pGAL1-BAX integrated into the genome and carried an empty vector. For the assay colonies were picked and diluted in sterile water. Using the comparative galactose induction assay as a template, the ODs at 600 nm were equalized to 0.035 and different dilutions of the colonies (10-0 to 10-3) were spotted on plates with glu-U-W agar and raff-U-W agar with 1% galactose in volumes of 10 µl. A yeast strain containing only pGAL1-BAX and the empty pWUS plasmid served as control (OV20) and was spotted along with the sample (Figure 2).
Figure 2: BAX rescue assay | In this assay, the strains OV19 and OV20 were grown on both glu-U-W agar (left) and raff-U-W (1% galactose) agar in different dilutions of OD600nm (10-0 to 10-3).
As BAX was expressed under the inducible promoter pGAL1 in both strains, we expected normal growth on the glu-U-W plates. On the raff-U-W plates with 1% galactose, we expected the control strain OV20 to show decreased colony size and number compared to the OV19 strain that should be rescued by the BI-1 plasmid. However, as seen on figure 2, no significant growth was seen for either strain in the presence of galactose. As such, we saw no indication that BI-1 was able to prevent BAX induced apoptosis. While it is possible that the lack of function of BI-1 could be the result of a loss of function during transformation, our data suggests BI-1 is not able to prevent BAX induced apoptosis in our system, and is therefore not suitable for our kill switch.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 954
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 377
- 1000COMPATIBLE WITH RFC[1000]