Difference between revisions of "Part:BBa K2365518:Experience"
Hitesh Gelli (Talk | contribs) (→Applications of BBa_K2365518) |
|||
| Line 5: | Line 5: | ||
===Applications of BBa_K2365518=== | ===Applications of BBa_K2365518=== | ||
| + | __NOTOC__ | ||
| + | <partinfo>BBa_K3190601 short</partinfo> | ||
| + | |||
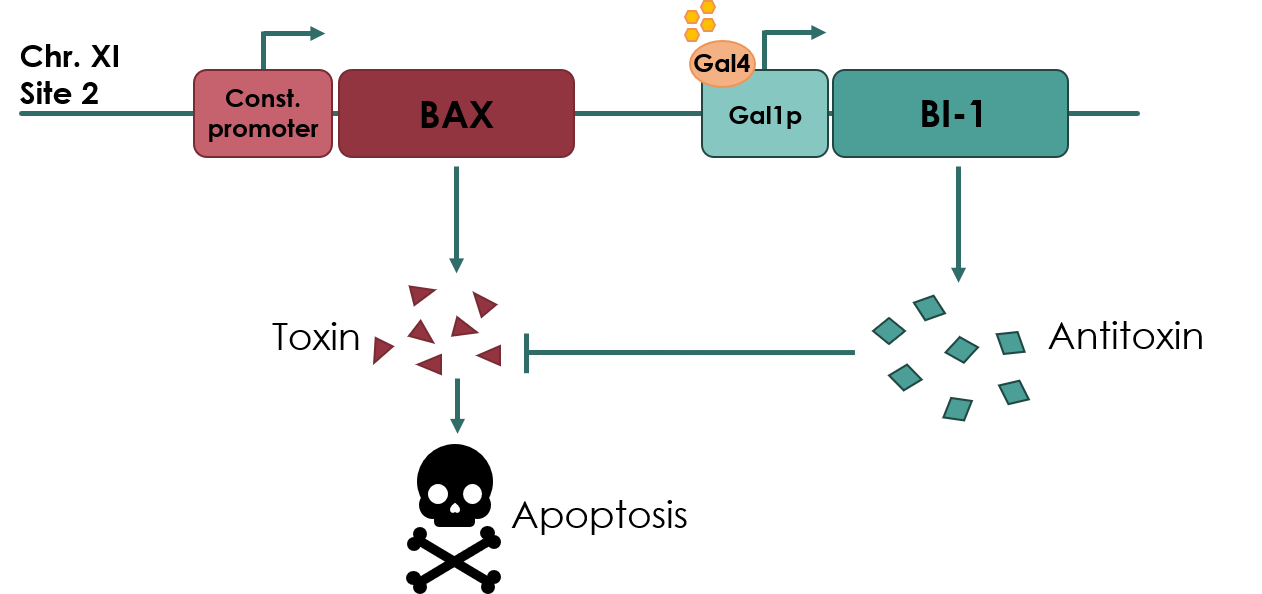
| + | Coding sequence of BAX Inhibitor-I (BI-1) anti-toxin under control of the galactose inducible GAL1 promoter (<partinfo>BBa_K3190050</partinfo>). This biobrick was designed for use in a yeast kill switch system, as described by NAU_CHINA 2017 (Figure 1). | ||
| + | |||
| + | ===Usage and Biology=== | ||
| + | <b><font size="3">Improvement of kill switch system</font></b> | ||
| + | |||
| + | We wanted to improve the BI-1 coding sequence (<partinfo>BBa_K2365518</partinfo>), which was submitted by the iGEM team NAU_CHINA 2017. They used it as part of their kill switch system and also submitted a composite part consisting of the TEF1 constitutive promoter, BI-1, and the yeast CYC1 terminator. | ||
| + | |||
| + | The kill switch proposed by team NAU China would work by co-expressing the BAX gene (<partinfo>BBa_K2365048</partinfo>), which encodes the pro-apoptotic Bax protein, under a constitutive promoter, together with BI-1 under the galactose inducible GAL1 promoter. In theory, as long as galactose is present in the media, BI-1 will be expressed, inhibiting the Bax protein. Should the yeast cell escape the media or matrix, BAX will be expressed, causing apoptosis of the cell. | ||
| + | |||
| + | [[File:UCopenhagen Killswitch.jpeg|600px]] | ||
| + | |||
| + | <small><b>Figure 1: Overview of the kill switch </b></small> | ||
| + | |||
| + | We improved the existing BI-1 sequence by expressing it under the control of the inducible pGAL1 promoter. This improvement will allow us to control the expression of BI-1, as the GAL1 promoter allows transcription of BI-1 only in the presence of galactose. | ||
| + | |||
| + | |||
| + | <b><font size="3">BAX rescue assay</font></b> | ||
| + | |||
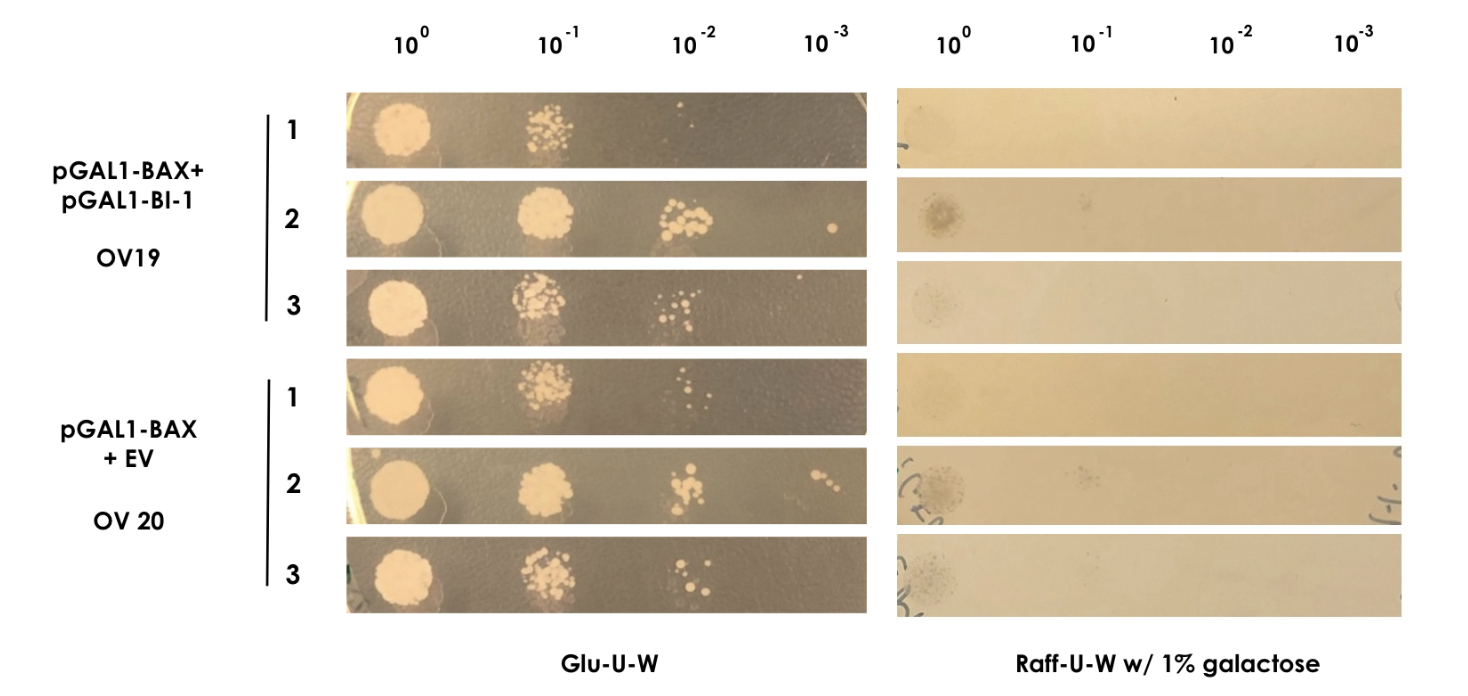
| + | In order to show that BI-1 is able to neutralize the lethal impact of BAX, we performed a rescue assay in which both BAX and BI-1 were placed under the control of the GAL1 promoter. For this, we constructed two strains; OV19, which had pGAL1-BAX integrated into the genome and pGAL1-BI-1 on a high-copy number episomal plasmid, and OV20, which had pGAL1-BAX integrated into the genome and carried an empty vector. | ||
| + | For the assay colonies were picked and diluted in sterile water. Using the comparative galactose induction assay as a template, the ODs at 600 nm were equalized to 0.035 and different dilutions of the colonies (10^-0 to 10^-3) were spotted on plates with glu-U-W agar and raff-U-W agar with 1% galactose in volumes of 10 µl. A yeast strain containing only pGAL1-BAX and the empty pWUS plasmid served as control (OV20) and was spotted along with the sample (Figure 2). | ||
| + | |||
| + | |||
| + | [[File:ovulaid35.png|800px]] | ||
| + | |||
| + | <small><b>Figure 2: BAX rescue assay</b> | In this assay, the strains OV19 and OV20 were grown on both glu-U-W agar (left) and raff-U-W (1% galactose) agar in different dilutions of OD600nm (10-0 to 10-3). </small> | ||
| + | |||
| + | As BAX was expressed under the inducible promoter pGAL1 in both strains, we expected normal growth on the glu-U-W plates. On the raff-U-W plates with 1% galactose, we expected the control strain OV20 to show decreased colony size and number compared to the OV19 strain that should be rescued by the BI-1 plasmid. However, as seen on figure 2, no significant growth was seen for either strain in the presence of galactose. As such, we saw no indication that BI-1 was able to prevent BAX induced apoptosis. While it is possible that the lack of function of BI-1 could be the result of a loss of function during transformation, our data suggests BI-1 is not able to prevent BAX induced apoptosis in our system, and is therefore not suitable for our kill switch. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K3190601 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | <!-- Uncomment this to enable Functional Parameter display | ||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K3190601 parameters</partinfo> | ||
| + | <!-- --> | ||
===User Reviews=== | ===User Reviews=== | ||
Revision as of 00:26, 22 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K2365518
pGAL1-BI-I: BI-I CDS under inducible promoter
Coding sequence of BAX Inhibitor-I (BI-1) anti-toxin under control of the galactose inducible GAL1 promoter (BBa_K3190050). This biobrick was designed for use in a yeast kill switch system, as described by NAU_CHINA 2017 (Figure 1).
Usage and Biology
Improvement of kill switch system
We wanted to improve the BI-1 coding sequence (BBa_K2365518), which was submitted by the iGEM team NAU_CHINA 2017. They used it as part of their kill switch system and also submitted a composite part consisting of the TEF1 constitutive promoter, BI-1, and the yeast CYC1 terminator.
The kill switch proposed by team NAU China would work by co-expressing the BAX gene (BBa_K2365048), which encodes the pro-apoptotic Bax protein, under a constitutive promoter, together with BI-1 under the galactose inducible GAL1 promoter. In theory, as long as galactose is present in the media, BI-1 will be expressed, inhibiting the Bax protein. Should the yeast cell escape the media or matrix, BAX will be expressed, causing apoptosis of the cell.
Figure 1: Overview of the kill switch
We improved the existing BI-1 sequence by expressing it under the control of the inducible pGAL1 promoter. This improvement will allow us to control the expression of BI-1, as the GAL1 promoter allows transcription of BI-1 only in the presence of galactose.
BAX rescue assay
In order to show that BI-1 is able to neutralize the lethal impact of BAX, we performed a rescue assay in which both BAX and BI-1 were placed under the control of the GAL1 promoter. For this, we constructed two strains; OV19, which had pGAL1-BAX integrated into the genome and pGAL1-BI-1 on a high-copy number episomal plasmid, and OV20, which had pGAL1-BAX integrated into the genome and carried an empty vector. For the assay colonies were picked and diluted in sterile water. Using the comparative galactose induction assay as a template, the ODs at 600 nm were equalized to 0.035 and different dilutions of the colonies (10^-0 to 10^-3) were spotted on plates with glu-U-W agar and raff-U-W agar with 1% galactose in volumes of 10 µl. A yeast strain containing only pGAL1-BAX and the empty pWUS plasmid served as control (OV20) and was spotted along with the sample (Figure 2).
Figure 2: BAX rescue assay | In this assay, the strains OV19 and OV20 were grown on both glu-U-W agar (left) and raff-U-W (1% galactose) agar in different dilutions of OD600nm (10-0 to 10-3).
As BAX was expressed under the inducible promoter pGAL1 in both strains, we expected normal growth on the glu-U-W plates. On the raff-U-W plates with 1% galactose, we expected the control strain OV20 to show decreased colony size and number compared to the OV19 strain that should be rescued by the BI-1 plasmid. However, as seen on figure 2, no significant growth was seen for either strain in the presence of galactose. As such, we saw no indication that BI-1 was able to prevent BAX induced apoptosis. While it is possible that the lack of function of BI-1 could be the result of a loss of function during transformation, our data suggests BI-1 is not able to prevent BAX induced apoptosis in our system, and is therefore not suitable for our kill switch.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 954
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 377
- 1000COMPATIBLE WITH RFC[1000]
User Reviews
UNIQfe9b58dbd3b81f9e-partinfo-00000005-QINU UNIQfe9b58dbd3b81f9e-partinfo-00000006-QINU