Difference between revisions of "Part:BBa K1614017"
Veronica Liu (Talk | contribs) |
Veronica Liu (Talk | contribs) |
||
| (10 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
{{Template:ssDNA}}<br/> | {{Template:ssDNA}}<br/> | ||
Aptamer binding kanamycin created by the MAWS (http://2015.igem.org/Team:Heidelberg/software/maws) Software from iGEM Team Heidelberg. | Aptamer binding kanamycin created by the MAWS (http://2015.igem.org/Team:Heidelberg/software/maws) Software from iGEM Team Heidelberg. | ||
| + | |||
| + | ===Abstarct=== | ||
| + | This kanamycin aptamer created by MAWS is characterized by DUT China A (https://2019.igem.org/Team:DUT_China_A/Characterization) in 2019. A method ,letting gold nanoparticles (Au NPs) and aptamers completely binds to the kanamycin ,was adopted to confirm that there is affinity activity between kanamycin and this aptamer. | ||
| + | |||
| + | ===Usage and Biology=== | ||
| + | This is an oligonucleotide with a seven-base sequence created by MAWS, which can bind to kanamycin. Depending on this property, it can be expected to be applied to detection of kanamycin. DUT China A 2019 provides a method to characterize this aptamer. The result indicates that this aptamer can bind to kanamycin as well as be used to detect kanamycin in a certain range of concentration. | ||
<h3>Characterization of this part by Gold nanoparticle-based colorimetric detection</h3> | <h3>Characterization of this part by Gold nanoparticle-based colorimetric detection</h3> | ||
| − | Team DUT China A 2019 characterized this part to find out whether this part can bind to kanamycin, we put forward a plan, Gold nanoparticle-based colorimetric detection. <br/> | + | Team DUT China A 2019(https://2019.igem.org/Team:DUT_China_A/Characterization) characterized this part to find out whether this part can bind to kanamycin, we put forward a plan, Gold nanoparticle-based colorimetric detection. <br/> |
<h4>Principle</h4> | <h4>Principle</h4> | ||
| − | Gold nanoparticles (Au NPs) covered by Citrate ion are charged negatively and they repel each other, remaining dispersive in the solution. After adding NaCl into this solution system, the double electrode layer of the Au NPs are destroyed and they tend to being coagulated, resulting the color of the solution changes from red to blue-purple, and the spectrum of the solution shifts red. The absorbance of 620nm obviously increase. However, short ssDNAs are readily adsorbed onto the surface of AuNPs without any modification and that such ssDNA-treated AuNPs are more stable than untreated ones. Adding aptamers into the system, the citrate root of nano gold surface is replaced. AuNPs can still remain negetively charged instead of being coagulated even if there is NaCl due to the non-specific interaction between DNA side chain phosphate groups and Au NPs, keeping the solution color red. When kanamycin, the determinand is added in to the solution, the adaptor configuration changes and falls off the surface of the Au NPs because there is | + | Gold nanoparticles (Au NPs) covered by Citrate ion are charged negatively and they repel each other, remaining dispersive in the solution. After adding NaCl into this solution system, the double electrode layer of the Au NPs are destroyed and they tend to being coagulated, resulting the color of the solution changes from red to blue-purple, and the spectrum of the solution shifts red. The absorbance of 620nm obviously increase. However, short ssDNAs are readily adsorbed onto the surface of AuNPs without any modification and that such ssDNA-treated AuNPs are more stable than untreated ones. Adding aptamers into the system, the citrate root of nano gold surface is replaced. AuNPs can still remain negetively charged instead of being coagulated even if there is NaCl due to the non-specific interaction between DNA side chain phosphate groups and Au NPs, keeping the solution color red. When kanamycin, the determinand is added in to the solution, the adaptor configuration changes and falls off the surface of the Au NPs because there is affinity between the kanamycn and the aptamers. AuNPs is going to coagulate. The changing concentrations of kanamycin lead to the changing AuNPs solution spectra. Adopting this method, the binding of the aptamer to kanamycin can be tested and the kanamycin in the solution can be easily and quickly detected. |
| Line 19: | Line 25: | ||
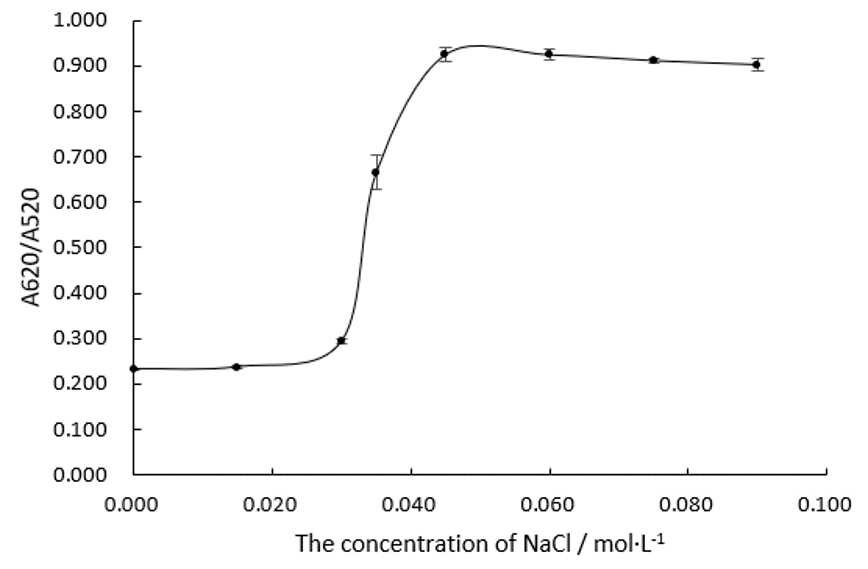
We fabricate Au NPs systems with different concentration of NaCl in 96 well plates and measure their absorbance ratios at wavelengths of 520 nm and 620 nm, which is shown in Figure.1 and Figure.2.<br/> | We fabricate Au NPs systems with different concentration of NaCl in 96 well plates and measure their absorbance ratios at wavelengths of 520 nm and 620 nm, which is shown in Figure.1 and Figure.2.<br/> | ||
| − | [[File:T--DUT China A--parts- | + | [[File:T--DUT China A--parts-NaClexplration.jpg|350px|thumb|Figure.1 A620/A520 of Au NPs systems determined by the concentration of NaCl]] |
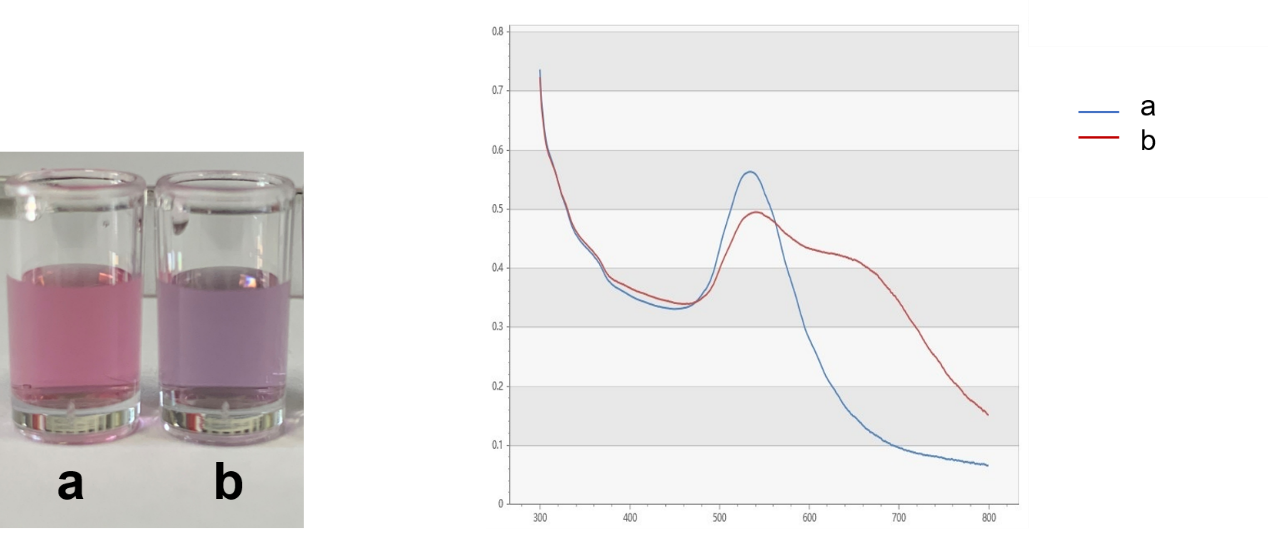
| − | [[File:T--DUT China A--parts- | + | [[File:T--DUT China A--parts-NaCl with and without.jpg|350px|thumb|Figure.2 Colors of AuNPs (150 nM) without NaCl (a) and with 50 nM of NaCl (b). The right graph shows absorption spectra of a and b.]] |
Obviously, the trend line of A620/A520 reaches a plateau when the concentration of NaCl comes to 0.045 M. We can assume that 0.045 M of NaCl is able to precipitate all the Au NPs in this system. | Obviously, the trend line of A620/A520 reaches a plateau when the concentration of NaCl comes to 0.045 M. We can assume that 0.045 M of NaCl is able to precipitate all the Au NPs in this system. | ||
| Line 29: | Line 35: | ||
Once there are extra aptamers in the solution system, they will bond to the kanamycin while the Au NPs are still totally protected, which will not lead to a change on A620/A520. We don’t want this happen. Therefore, we must ensure that the aptamers at the working concentration are not excessive. | Once there are extra aptamers in the solution system, they will bond to the kanamycin while the Au NPs are still totally protected, which will not lead to a change on A620/A520. We don’t want this happen. Therefore, we must ensure that the aptamers at the working concentration are not excessive. | ||
We fabricate Au NPs systems with different concentration of Aptamer K3 in 96 well plates when the NaCl is 0.075 M and measure their absorbance ratios at wavelengths of 520 nm and 620 nm, which is shown in Figure.3. | We fabricate Au NPs systems with different concentration of Aptamer K3 in 96 well plates when the NaCl is 0.075 M and measure their absorbance ratios at wavelengths of 520 nm and 620 nm, which is shown in Figure.3. | ||
| − | [[File:T--DUT China A--parts- | + | [[File:T--DUT China A--parts-k3-figure3.jpg|350px|thumb|Figure.3 A620/A520 of Au NPs systems determined by the concentration of Aptamer K3. The concentration of Au NPs is 150 nM. The concentration of NaCl is 0.075 M.]] |
| − | Obviously, the trend line of A620/A520 is likely to just reach a plateau when the concentration of Aptamer K3 comes to 0.20 μM. We can assume that | + | Obviously, the trend line of A620/A520 is likely to just reach a plateau when the concentration of Aptamer K3 comes to 0.20 μM. We can assume that Aptamer K3 at this amount are just enough to protect all the Au NPs at 150 nM. |
In the following experiment 4, we will make the concentration of Aptamer at 0.20 μM in the system. | In the following experiment 4, we will make the concentration of Aptamer at 0.20 μM in the system. | ||
| − | <strong>Experiment 4: Characterization of | + | <strong>Experiment 4: Characterization of Aptamer K3</strong><br/> |
Incubate 150 nM of AuNP and 0.20μM of Aptamer K3 for 30min at 37℃, with system volume being 200 μL. Add different concentrations of kanamycin into the system with the final concentration of kanamycin between 0 and 0.45 μM. | Incubate 150 nM of AuNP and 0.20μM of Aptamer K3 for 30min at 37℃, with system volume being 200 μL. Add different concentrations of kanamycin into the system with the final concentration of kanamycin between 0 and 0.45 μM. | ||
Add 5μL, 3M of NaCl solution, making its final concentration at 0.075 M. All these steps are done in 96-well plates. Measure the absorbance at 620nm and 520nm by uv-vis spectrometer, and the changes of A 620 / A 520 are recorded and analyzed, which is shown in Figure.4. | Add 5μL, 3M of NaCl solution, making its final concentration at 0.075 M. All these steps are done in 96-well plates. Measure the absorbance at 620nm and 520nm by uv-vis spectrometer, and the changes of A 620 / A 520 are recorded and analyzed, which is shown in Figure.4. | ||
| − | [[File:T--DUT China A--parts- | + | [[File:T--DUT China A--parts-k3-figure4.jpg|350px|thumb|Figure.4 A620/A520 of Au NPs systems determined by the concentration of Kanamycin. The concentration of Au NPs is 150 nM. The concentration of NaCl is 0.075 M. The concentration of Aptamer K3 is 0.20 μM.]] |
| + | |||
| + | <h4>Conclusion</h4> | ||
| + | As the trend line in Figure.4 shows, A620/A520 changes as the concentration of kanamycin changes and finally reaches a plateau, indicating that the changing concentrations of kanamycin lead to the changing AuNPs solution spectra. We can make a conclusion that there is binding between Aptamer K3 and kanamycin.<br/> | ||
| + | |||
| + | |||
| + | ===Reference=== | ||
| + | [1] Song K M, Cho M, Jo H, et al. Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer[J]. Analytical biochemistry, 2011, 415(2): 175-181.<br/> | ||
| + | [2] Storhoff J J, Elghanian R, Mucic R C, et al. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes[J]. Journal of the American Chemical Society, 1998, 120(9): 1959-1964. | ||
| − | |||
| − | |||
<br/> | <br/> | ||
<br/> | <br/> | ||
| Line 47: | Line 59: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| − | + | <br/> | |
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
| + | <br/> | ||
<!-- --> | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
Latest revision as of 22:58, 21 October 2019
software generated kanamycin aptamer (candidate 3)
Notice: Functional DNA
This part is a sequence of a functional ssDNA. It is only active as single-stranded DNA. It can not be cloned into a plasmid. For use order it as a DNA oligo.
Aptamer binding kanamycin created by the MAWS (http://2015.igem.org/Team:Heidelberg/software/maws) Software from iGEM Team Heidelberg.
Abstarct
This kanamycin aptamer created by MAWS is characterized by DUT China A (https://2019.igem.org/Team:DUT_China_A/Characterization) in 2019. A method ,letting gold nanoparticles (Au NPs) and aptamers completely binds to the kanamycin ,was adopted to confirm that there is affinity activity between kanamycin and this aptamer.
Usage and Biology
This is an oligonucleotide with a seven-base sequence created by MAWS, which can bind to kanamycin. Depending on this property, it can be expected to be applied to detection of kanamycin. DUT China A 2019 provides a method to characterize this aptamer. The result indicates that this aptamer can bind to kanamycin as well as be used to detect kanamycin in a certain range of concentration.
Characterization of this part by Gold nanoparticle-based colorimetric detection
Team DUT China A 2019(https://2019.igem.org/Team:DUT_China_A/Characterization) characterized this part to find out whether this part can bind to kanamycin, we put forward a plan, Gold nanoparticle-based colorimetric detection.
Principle
Gold nanoparticles (Au NPs) covered by Citrate ion are charged negatively and they repel each other, remaining dispersive in the solution. After adding NaCl into this solution system, the double electrode layer of the Au NPs are destroyed and they tend to being coagulated, resulting the color of the solution changes from red to blue-purple, and the spectrum of the solution shifts red. The absorbance of 620nm obviously increase. However, short ssDNAs are readily adsorbed onto the surface of AuNPs without any modification and that such ssDNA-treated AuNPs are more stable than untreated ones. Adding aptamers into the system, the citrate root of nano gold surface is replaced. AuNPs can still remain negetively charged instead of being coagulated even if there is NaCl due to the non-specific interaction between DNA side chain phosphate groups and Au NPs, keeping the solution color red. When kanamycin, the determinand is added in to the solution, the adaptor configuration changes and falls off the surface of the Au NPs because there is affinity between the kanamycn and the aptamers. AuNPs is going to coagulate. The changing concentrations of kanamycin lead to the changing AuNPs solution spectra. Adopting this method, the binding of the aptamer to kanamycin can be tested and the kanamycin in the solution can be easily and quickly detected.
Protocol, Results and Discussion
Experiment 1: Preparation for Au NPs
Add 125mL HAuCl(1mM) water solution to 250mL round bottom flask. Heat it until boils then quickly add 12.5 mL (38.8mM) sodium citrate solution into this system. Heat for 10min, remove the heating sleeve, stir for 15min. Cool to room temperature and obtain Au NPs with particle size of about 13nm.
Experiment 2: Exploration on the least concentration of NaCl to precipitate a certain concentration of Au NPs
We fabricate Au NPs systems with different concentration of NaCl in 96 well plates and measure their absorbance ratios at wavelengths of 520 nm and 620 nm, which is shown in Figure.1 and Figure.2.
Obviously, the trend line of A620/A520 reaches a plateau when the concentration of NaCl comes to 0.045 M. We can assume that 0.045 M of NaCl is able to precipitate all the Au NPs in this system. In the following experiments, we will make the concentration of NaCl at 0.075 M in every system.
Experiment 3: Explorations on the least concentration of Aptamer K3 to protect all the Au NPs from being precipitated
Once there are extra aptamers in the solution system, they will bond to the kanamycin while the Au NPs are still totally protected, which will not lead to a change on A620/A520. We don’t want this happen. Therefore, we must ensure that the aptamers at the working concentration are not excessive.
We fabricate Au NPs systems with different concentration of Aptamer K3 in 96 well plates when the NaCl is 0.075 M and measure their absorbance ratios at wavelengths of 520 nm and 620 nm, which is shown in Figure.3.
Obviously, the trend line of A620/A520 is likely to just reach a plateau when the concentration of Aptamer K3 comes to 0.20 μM. We can assume that Aptamer K3 at this amount are just enough to protect all the Au NPs at 150 nM. In the following experiment 4, we will make the concentration of Aptamer at 0.20 μM in the system.
Experiment 4: Characterization of Aptamer K3
Incubate 150 nM of AuNP and 0.20μM of Aptamer K3 for 30min at 37℃, with system volume being 200 μL. Add different concentrations of kanamycin into the system with the final concentration of kanamycin between 0 and 0.45 μM.
Add 5μL, 3M of NaCl solution, making its final concentration at 0.075 M. All these steps are done in 96-well plates. Measure the absorbance at 620nm and 520nm by uv-vis spectrometer, and the changes of A 620 / A 520 are recorded and analyzed, which is shown in Figure.4.
Conclusion
As the trend line in Figure.4 shows, A620/A520 changes as the concentration of kanamycin changes and finally reaches a plateau, indicating that the changing concentrations of kanamycin lead to the changing AuNPs solution spectra. We can make a conclusion that there is binding between Aptamer K3 and kanamycin.
Reference
[1] Song K M, Cho M, Jo H, et al. Gold nanoparticle-based colorimetric detection of kanamycin using a DNA aptamer[J]. Analytical biochemistry, 2011, 415(2): 175-181.
[2] Storhoff J J, Elghanian R, Mucic R C, et al. One-pot colorimetric differentiation of polynucleotides with single base imperfections using gold nanoparticle probes[J]. Journal of the American Chemical Society, 1998, 120(9): 1959-1964.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]