Difference between revisions of "Part:BBa K3081055"
| Line 18: | Line 18: | ||
https://2019.igem.org/wiki/images/6/6a/T--Peking--picture_for_ssrA_new.jpeg | https://2019.igem.org/wiki/images/6/6a/T--Peking--picture_for_ssrA_new.jpeg | ||
| − | Figure 1: Characterization of CRISPRri-ssrA system. (A) Comparison between dCas9 and dCas9-ssrA system by expression level and CRISPRi effect on mRFP fluorescence. (B) Comparison of effect on cell growth between CRISPRri and CRISPRri-ssrA, both targeted to R1+ box(left:R1+; right:R1+ ssrA). (C) Reversibility of CRISPRri-ssrA system targeted to R1+ box. Hollow arrows stand for removal of arabinose while solid black arrows stand for addition of arabinose. | + | Figure 1: Characterization of CRISPRri-ssrA system. (A) Comparison between dCas9 and dCas9-ssrA system by expression level and CRISPRi effect on mRFP fluorescence. (B) Comparison of effect on cell growth between CRISPRri and CRISPRri-ssrA, both targeted to R1+ box (left:R1+; right:R1+ ssrA). (C) Reversibility of CRISPRri-ssrA system targeted to R1+ box. Hollow arrows stand for removal of arabinose while solid black arrows stand for addition of arabinose. |
Reference: | Reference: | ||
Revision as of 20:03, 21 October 2019
dCas9-ssrA: a fast degradation tag "ssrA" linked to the C-terminal of dCas9
In our experiment, we found that the inhibitory effect of dCas9 to DNA replication shows to have over-inhibition on cell growth and small dynamic range when the sgRNAs targeting high affinity DnaA boxes. In order to extend this system to combine all DnaA boxes, we improve the performance of the CRISPRri system to weaken its effect by adding a degradation signal peptide ssrA to dCas9. Thus, we get an improved part (BBa_K3081055) from dCas9 (BBa_K1150000).
The ssrA peptide tag from Mycoplasma florum has been developed as a versatile biotechnology tool to control orthogonal degradation of tagged proteins in Escherichia coli [1]. Ideally, after fused dCas9 and ssrA, it accelerates the degradation rate of dCas9 and thus weaken the inhibition of DNA replication by CRISPRri.
As a preliminary verification, firstly, we fused ssrA tag to dCas9-sfGFP, and we tested the expression curve of dCas9-sfGFP-ssrA by adding different concentration of inducer (Figure 1A). We found that the protein with ssrA was expressed at a lower level than the control protein, with the same concentration of inducer. We believe that because ssrA mediates protein degradation, the equilibrium concentration of the protein is reduced.
To test the function of fused version of dCas9-ssrA, we co-transformed two plasmids to one E. coli cell, which express mRFP and dCas9-ssrA with sgRNA targeting CDS of mRFP, respectively. As a matter of fact, the new CRISPRi system shows a gentler decrease in fluorescence when dCas9 is fused with ssrA tag, while non-binding dCas9 with or without ssrA has no influence on mRFP expression (Figure 1B). This result confirmed fusion with ssrA, the original function of dCas9 is unaffected.
So, we tested the improved CRISPRri-ssrA system with targeting site of DnaA boxes which are shown to have excessive inhibition on cell growth before. We found that the degradation tag makes inhibition effect much milder, which allows for a wider adjusting range (Figure 1C).
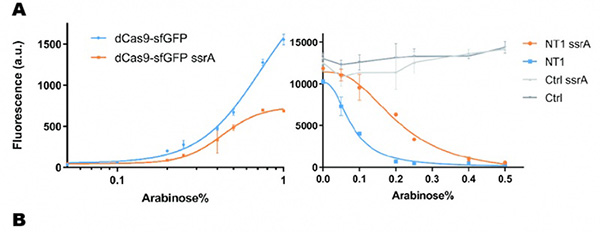


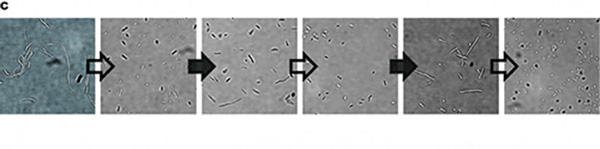
Figure 1: Characterization of CRISPRri-ssrA system. (A) Comparison between dCas9 and dCas9-ssrA system by expression level and CRISPRi effect on mRFP fluorescence. (B) Comparison of effect on cell growth between CRISPRri and CRISPRri-ssrA, both targeted to R1+ box (left:R1+; right:R1+ ssrA). (C) Reversibility of CRISPRri-ssrA system targeted to R1+ box. Hollow arrows stand for removal of arabinose while solid black arrows stand for addition of arabinose.
Reference:
[1] Lv, L., Wu, Y., Zhao, G., & Qi, H. (2019). Improvement in the Orthogonal Protein Degradation in Escherichia coli by Truncated mf-ssrA Tag. Transactions of Tianjin University, 1-7.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 1099
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 3378
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
