Difference between revisions of "Part:BBa K3016100"
| (11 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
| − | This part contains <i>Vibrio natriegens'</i> TorA Tat signal peptide. It is a twin-arginine (RR) motif containing signal peptide for periplasmic transport of proteins via the twin-arginine translocation (Tat) pathway. Derived from Vibrio natriegens’ <i>torA | + | This part contains <i>Vibrio natriegens'</i> TorA Tat signal peptide. It is a twin-arginine (RR) motif containing signal peptide for periplasmic transport of proteins via the twin-arginine translocation (Tat) pathway. Derived from Vibrio natriegens’ TMAO reductase (<i>torA</i>) gene. |
| + | Note: TorA signal peptide may be prone to inclusion body formation in <i>Escherichia coli</i> (Jong <i>et al.</i>, 2017). Unconfirmed in <i>Vibrio natriegens</i>. | ||
| − | + | ===Biology=== | |
| − | ==Biology== | + | |
The twin-arginine translocation (Tat) pathway is capable of translocating fully folded proteins up to 150 kDa. It also contains a quality control feature of rejecting misfolded proteins. In some cases, disulfide bridge formation is not required for successful translocation. (Alanen et al., 2015) | The twin-arginine translocation (Tat) pathway is capable of translocating fully folded proteins up to 150 kDa. It also contains a quality control feature of rejecting misfolded proteins. In some cases, disulfide bridge formation is not required for successful translocation. (Alanen et al., 2015) | ||
| Line 18: | Line 18: | ||
This part can be N-terminally fused with your protein of choice to facilitate its periplasmic transport via the twin-arginine translocation (Tat) pathway. | This part can be N-terminally fused with your protein of choice to facilitate its periplasmic transport via the twin-arginine translocation (Tat) pathway. | ||
| − | Aalto-Helsinki 2019 | + | |
| + | |||
| + | ==Characterization== | ||
| + | |||
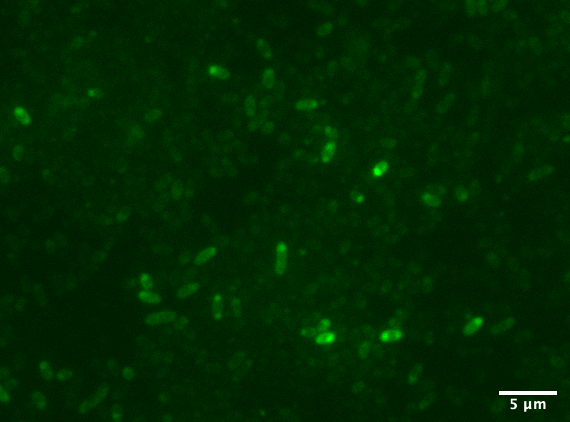
| + | Aalto-Helsinki 2019 characterized this part by combining it with YGFP ([https://parts.igem.org/Part:BBa_K3016600 BBa_K3016600]), a slow-bleaching GFP variant, creating a composite TorA-YGFP part ([https://parts.igem.org/Part:BBa_K3016200 BBa_K3016200]) to test protein translocation into <i>Vibrio natriegens'</i> and <i>Escherichia coli</i> DH5a's periplasm. | ||
| + | |||
| + | [[Image:T--Aalto-Helsinki--TorA-YGFP_natriegens.png|thumb|530px|center|<font size="1">Vibrio natriegens cells expressing TorA-YGFP</font>]] | ||
| + | |||
| + | |||
| + | [[Image:T--Aalto-Helsinki--TorA-YGFP_coli.jpg|thumb|530px|center|<font size="1"><i>Escherichia coli</i> DH5a cells expressing TorA-YGFP</font>]] | ||
| + | |||
| + | |||
| + | In the images above we see <i>Vibrio natriegens</i> and <i>Escherichia coli</i> DH5a cells expressing TorA-YGFP. Note the polar localisation of fluorescence. This can be a sign of periplasmic localisation of YGFP under osmotic pressure (Sochacki <i>et al.</i>, 2011) or inclusion body formation (Jong <i>et al.</i>, 2017). | ||
| + | |||
| + | |||
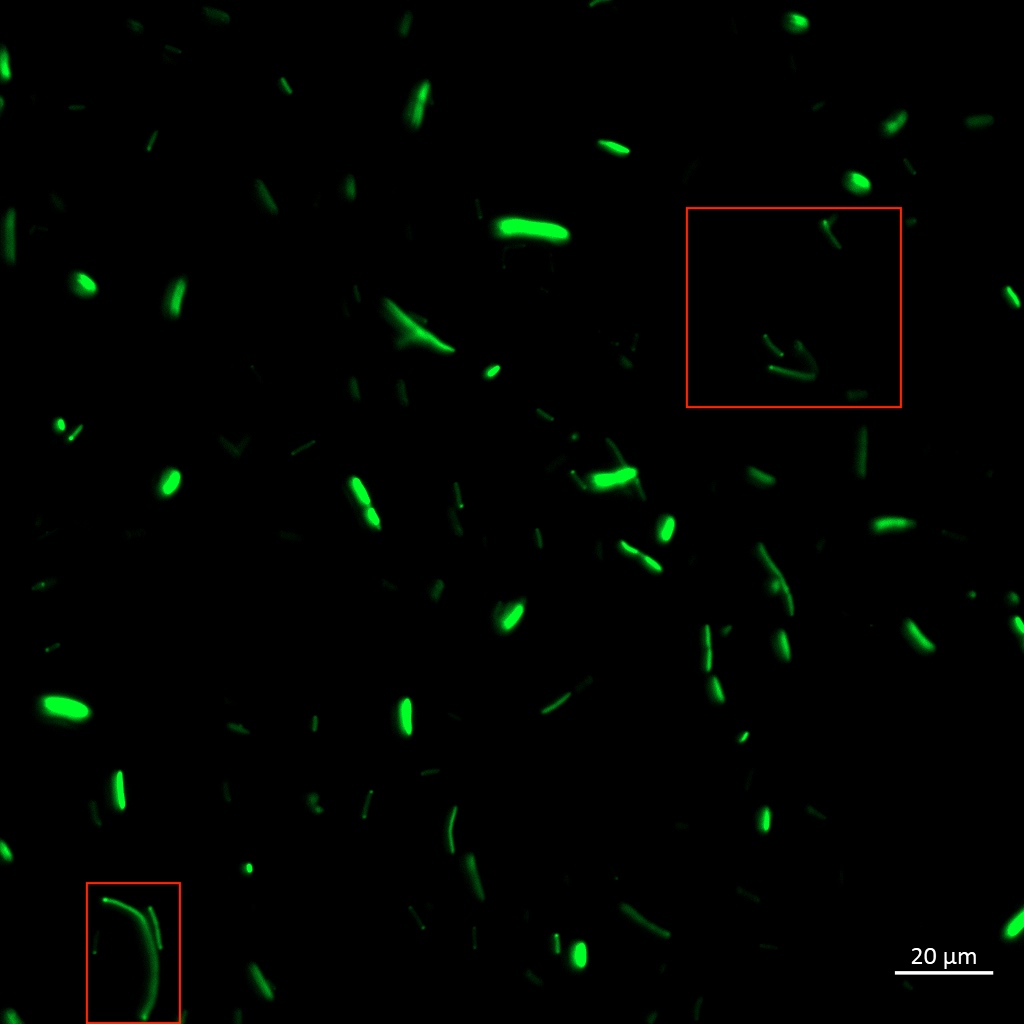
| + | To be certain of successful periplasmic translocation, a cell fractionation experiment was performed on <i>Vibrio natriegens</i>. The periplasmic fraction of TorA-YGFP expressing cells was extracted ([https://2019.igem.org/wiki/images/e/e4/T--Aalto-Helsinki--protocols_AH_new.pdf PureFrac-protocol]) and placed under UV light, image below. The presence of YGFP in the periplasmic fractions can be seen clearly. | ||
| + | |||
| + | |||
| + | [[Image:T--Aalto-Helsinki--TorA-YGFP_natriegens_periplasmic_fraction.jpg|thumb|530px|center|<font size="1">Periplasmic fractions of Vibrio natriegens cells expressing TorA-YGFP </font>]] | ||
| + | |||
| + | |||
| + | These results indicate that the <i>Vibrio natriegens'</i> TorA signal peptide successfully translocates proteins into the periplasm of <i>Vibrio natriegens</i>, and possibly even of <i>Escherichia coli</i>. | ||
| + | |||
<!-- --> | <!-- --> | ||
| Line 24: | Line 46: | ||
<partinfo>BBa_K3016100 SequenceAndFeatures</partinfo> | <partinfo>BBa_K3016100 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | ==References:== | ||
| + | |||
| + | Alanen, H. I., Walker, K. L., Suberbie, M. L. V., Matos, C. F., Bönisch, S., Freedman, R. B., ... & Robinson, C. (2015). Efficient export of human growth hormone, interferon α2b and antibody fragments to the periplasm by the Escherichia coli Tat pathway in the absence of prior disulfide bond formation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1853(3), 756-763. | ||
| + | |||
| + | Jong, W. S., Vikström, D., Houben, D., de Gier, J. W., & Luirink, J. (2017). Application of an E. coli signal sequence as a versatile inclusion body tag. Microbial cell factories, 16(1), 50. | ||
| + | |||
| + | Sochacki, K. A., Shkel, I. A., Record, M. T., & Weisshaar, J. C. (2011). Protein diffusion in the periplasm of E. coli under osmotic stress. Biophysical journal, 100(1), 22-31. | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 18:05, 21 October 2019
Vibrio natriegens' TorA signal peptide
This part contains Vibrio natriegens' TorA Tat signal peptide. It is a twin-arginine (RR) motif containing signal peptide for periplasmic transport of proteins via the twin-arginine translocation (Tat) pathway. Derived from Vibrio natriegens’ TMAO reductase (torA) gene.
Note: TorA signal peptide may be prone to inclusion body formation in Escherichia coli (Jong et al., 2017). Unconfirmed in Vibrio natriegens.
Biology
The twin-arginine translocation (Tat) pathway is capable of translocating fully folded proteins up to 150 kDa. It also contains a quality control feature of rejecting misfolded proteins. In some cases, disulfide bridge formation is not required for successful translocation. (Alanen et al., 2015)
Translocation using the tat-pathway requires the protein to contain a N-terminal signal peptide with a twin-arginine (RR) motif. The signal peptide is cleaved during the translocation process. A pair of V. natriegens’ native twin-arginine signal peptides identified by Aalto-Helsinki can be found here (TorAand Aminotransferase)
Use
This part can be N-terminally fused with your protein of choice to facilitate its periplasmic transport via the twin-arginine translocation (Tat) pathway.
Characterization
Aalto-Helsinki 2019 characterized this part by combining it with YGFP (BBa_K3016600), a slow-bleaching GFP variant, creating a composite TorA-YGFP part (BBa_K3016200) to test protein translocation into Vibrio natriegens' and Escherichia coli DH5a's periplasm.
In the images above we see Vibrio natriegens and Escherichia coli DH5a cells expressing TorA-YGFP. Note the polar localisation of fluorescence. This can be a sign of periplasmic localisation of YGFP under osmotic pressure (Sochacki et al., 2011) or inclusion body formation (Jong et al., 2017).
To be certain of successful periplasmic translocation, a cell fractionation experiment was performed on Vibrio natriegens. The periplasmic fraction of TorA-YGFP expressing cells was extracted (PureFrac-protocol) and placed under UV light, image below. The presence of YGFP in the periplasmic fractions can be seen clearly.
These results indicate that the Vibrio natriegens' TorA signal peptide successfully translocates proteins into the periplasm of Vibrio natriegens, and possibly even of Escherichia coli.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References:
Alanen, H. I., Walker, K. L., Suberbie, M. L. V., Matos, C. F., Bönisch, S., Freedman, R. B., ... & Robinson, C. (2015). Efficient export of human growth hormone, interferon α2b and antibody fragments to the periplasm by the Escherichia coli Tat pathway in the absence of prior disulfide bond formation. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1853(3), 756-763.
Jong, W. S., Vikström, D., Houben, D., de Gier, J. W., & Luirink, J. (2017). Application of an E. coli signal sequence as a versatile inclusion body tag. Microbial cell factories, 16(1), 50.
Sochacki, K. A., Shkel, I. A., Record, M. T., & Weisshaar, J. C. (2011). Protein diffusion in the periplasm of E. coli under osmotic stress. Biophysical journal, 100(1), 22-31.