Difference between revisions of "Part:BBa K3031018"
| (9 intermediate revisions by the same user not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K3031018 short</partinfo> | <partinfo>BBa_K3031018 short</partinfo> | ||
| + | |||
| + | |||
| + | OxyR is a Transcription factor protein. OxyR reacts with H2O2 resulting in the oxidation of a reactive cysteine (Cys-199) formation of a intermolecular disulfide bond with neighboring cyctein-208 and thus a conformation change in the OxyR protein occurs. This change is shape activates the OxyR protein which then can bind to certain promoters with binding site regions to regulate the expression of downstream genes. | ||
<html> | <html> | ||
| − | |||
<figure class="figure-center"> | <figure class="figure-center"> | ||
<img src="https://2019.igem.org/wiki/images/3/36/T--SUIS_Shanghai--_OxyRdimer.png" align="center" alt="OxyR"> | <img src="https://2019.igem.org/wiki/images/3/36/T--SUIS_Shanghai--_OxyRdimer.png" align="center" alt="OxyR"> | ||
| Line 19: | Line 21: | ||
<figure class="figure-center"> | <figure class="figure-center"> | ||
<img src="https://2019.igem.org/wiki/images/5/53/T--SUIS_Shanghai--_PDBworkflow.png" height=400 width=600 align="center" alt="Cys_SOH"> | <img src="https://2019.igem.org/wiki/images/5/53/T--SUIS_Shanghai--_PDBworkflow.png" height=400 width=600 align="center" alt="Cys_SOH"> | ||
| − | <figcaption><b>Workflow outlining the selection criteria of all the 3D crystal protein structures used in this | + | <figcaption><b>Workflow outlining the selection criteria of all the 3D crystal protein structures used in this study to inform the improvement of the OxyR part</b>.</figcaption> |
</figure> | </figure> | ||
<p>The Amino acid, solvent, and ligand contacts for each atom contained within all Cys-SOH and unmodified cysteine sites in the dataset was furthered analyzed using PISA (Protein Interphases, Surfaces and Assemblies) (Krissinel & Henrick, 2007). The CCP4 Graphical User Interface (CCP4i) (Potterton, Briggs, Turkenburg, & Dodson, 2003) was used to perform and visualize PISA calculations. Crystal structures of protein containing sites of interest in PDB format were used as inputs into the interface. Default parameters were used and only calculations for contacts between 2.0 to 3.2 Angstroms within the chain containing the residue of interest were obtained. | <p>The Amino acid, solvent, and ligand contacts for each atom contained within all Cys-SOH and unmodified cysteine sites in the dataset was furthered analyzed using PISA (Protein Interphases, Surfaces and Assemblies) (Krissinel & Henrick, 2007). The CCP4 Graphical User Interface (CCP4i) (Potterton, Briggs, Turkenburg, & Dodson, 2003) was used to perform and visualize PISA calculations. Crystal structures of protein containing sites of interest in PDB format were used as inputs into the interface. Default parameters were used and only calculations for contacts between 2.0 to 3.2 Angstroms within the chain containing the residue of interest were obtained. | ||
| Line 25: | Line 27: | ||
<figure class="figure-center"> | <figure class="figure-center"> | ||
<img src="https://2019.igem.org/wiki/images/9/91/T--SUIS_Shanghai--AAmodified.png" height=250 width=300 align="left" alt="Cys_SOH"> | <img src="https://2019.igem.org/wiki/images/9/91/T--SUIS_Shanghai--AAmodified.png" height=250 width=300 align="left" alt="Cys_SOH"> | ||
| − | + | </figure> | |
| − | + | ||
<figure class="figure-center"> | <figure class="figure-center"> | ||
<img src="https://2019.igem.org/wiki/images/7/7a/T--SUIS_Shanghai--Unmod.png" height=250 width=300 align="right" alt="Cys_SOH"> | <img src="https://2019.igem.org/wiki/images/7/7a/T--SUIS_Shanghai--Unmod.png" height=250 width=300 align="right" alt="Cys_SOH"> | ||
| − | < | + | </figure> |
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <p><b>Amino Acid composition surrounding the 3.5A space around the S-sulfenylated cystienes (left) and th unmodified cysteines (right)</b></p> | ||
| + | <br> | ||
| + | <p>The solvent accessible surface area (SASA) of Cys-199 was determined using AREAMOL. Areaimol is a tool from the collection of programs from the Collaborative Computational Project No. 4 (CCP4) software suite version 7.0 (Winn et al., 2011). It finds the solvent accessible area of atoms in a PDB coordinate file by calculating the area of an atoms Van der Waals surface that a probe sphere rolls over (Lee & Richards, 1971). The default probe radius of 1.4 Angstroms was used to determine SASA. The CCP4 Graphical User Interface (CCP4i) (Potterton et al., 2003) was used to perform AREAIMOL calculations. </p> | ||
| + | <br> | ||
| + | <p><b>SASA of cysteine-199 in reduced form (PDB ID: 1i69) = 21.5 Angstroms</b><p> | ||
| + | <p>Relative solvent accessibility (RSA) was determined by dividing the SASA by the max accessible surface area (maxASA). | ||
| + | <br> | ||
| + | <ol> | ||
| + | <li>RSA = SASA/maxASA</li> | ||
| + | <li>where: RSA = relative Solvent Accessibility</li> | ||
| + | <li>SASA = Solvent Accessibly Surface Area</li> | ||
| + | <li>maxASA = maximum Accessible Surface Area</li></ol> | ||
| + | <br> | ||
| + | <p>The maxASA value used for cysteine 167.0 Angstroms (Tian et al., 2013). An RSA of 10% was used to be the cut off for classifying a residue as surface accessible (RSA > 10%) or buried (RSA < 10%). A comparison of modified and unmodified cysteine RSA values was conducted in order to determine if there was enrichment for exposed or buried surfaces within the Cys-SOH environments. A quantile– quantile probability plot (Q-Q plot) was used to determine if there was enrichment for exposed or buried cysteines within the Cys-SOH compared to unmodified sites. RSA values of modified and unmodified cysteine residues were sorted and values of RSA for each quantile was plotted against each other (i.e. RSA values of modified and unmodified cysteine residues were sorted from 0.01 to 1.0 and plotted against each other). </p> | ||
| + | <br> | ||
| + | <figure class="figure-center"> | ||
| + | <img src="https://2019.igem.org/wiki/images/6/61/T--SUIS_Shanghai--Quantile.png" height=300 width=400 align="center" alt="QQ"> | ||
| + | <figcaption><b>Quantile Quantile plot showing that S-sulfenylated cysteines have a higher distribution of relative solvent accessibility (RSA) values skewed towards a larger solvent accessibility (i.e. higher RSA) when compared to unmodified cysteines. This plot was constructed by sorting the RSA values of S-sulfenylated cysteines in our data set (n = 373) and unmodified reduced cysteine forms (n = 426) into percentiles and plotting them against each other. The diagonal line represents a line where all points would fall should the RSA values follow an equal distribution between sites.</b></figcaption> | ||
| + | </figure> | ||
| + | <br> | ||
| + | <p><b>Mutations made to OxyR (part BBa_K1104200):</b><p> | ||
| + | <ol> | ||
| + | <li><b>Residue 203: Glutamine (wild type) mutated to Serine</b> | ||
| + | Reasoning: Serine is overrepresented at position + 4 in the amino acid sequences flanking the S-sulfenylated cysteines (fig 7). In OxyR this position was filled with a Glutamine residue. We decided that placing a Serine there may allow for an improvement sensitivity to oxidation. From the literature accompanied by our modeling it was found that Serine, as a residue being an uncharged H-bond donor, likely has an important role in the activation of Cysteine, mainly by lowering its pKa (Mariano & Gladyshev, 2011). These two pieces of evidence allowed us to make this decision to mutate the protein.</li> | ||
| + | |||
| + | <li><b>Residue 149: Valine (wild type) mutated to Glycine</b> | ||
| + | |||
| + | We replaced on hydrophobic residue with another smaller sized one in the hope of increasing the solvent accessibility on the cysteine’s thiol. Val-149 was in close contact with Cys-199 as described below. </li></ol> | ||
| + | <br> | ||
| + | <figure class="figure-center"> | ||
| + | <img src="https://2019.igem.org/wiki/images/b/b2/T--SUIS_Shanghai--OxyR.png" height=400px width=500px align="center" alt="Original OxyR"> | ||
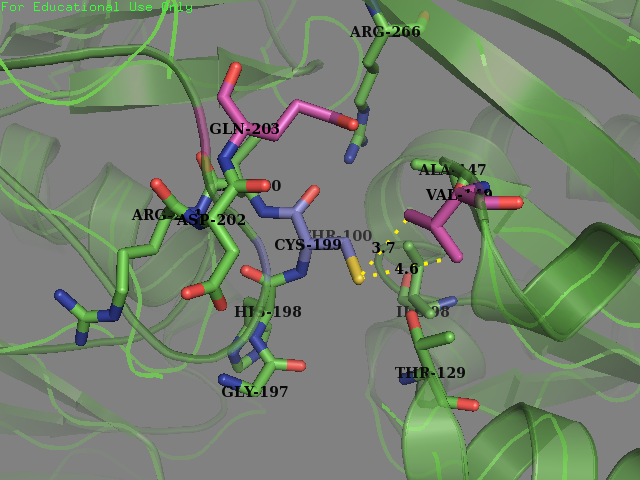
| + | <figcaption><b>Original OxyR Protein:</b>Environment surrounding the reactive cysteine-199 of the OxyR protein crystal structure protein (PDB ID: 1i69). Cysteine-199 is colored in Blue and the residues chosen for mutagenesis are highlighted in magenta (Val-147 and Glu-203).</figecaption> | ||
| + | </figure> | ||
| + | <figure class="figure-center"> | ||
| + | <img src="https://2019.igem.org/wiki/images/a/a2/T--SUIS_Shanghai--OxyRMutated.png" height=400px width=500px align="center" alt="Mutated OxyR"> | ||
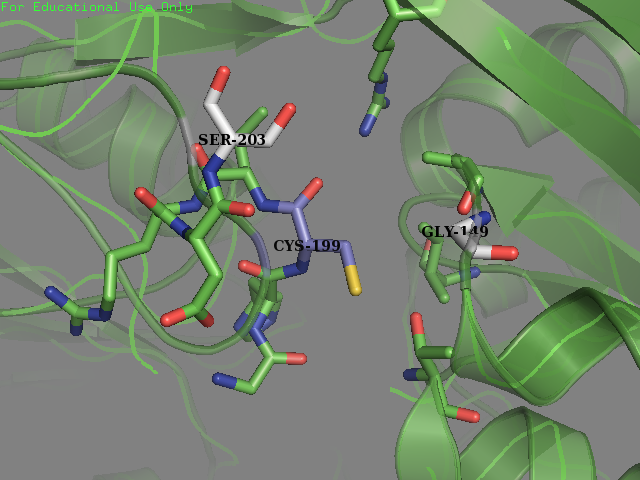
| + | <figcaption><b>Mutated OxyR:</b>Environment surrounding the reactive cysteine-199 of the OxyR protein crystal structure in reduced form (PDB ID: 1i69) after mutagenesis. New residues (Gly-147 and Ser-203) are colored in light grey.</figcaption> | ||
| + | </figure> | ||
| + | <br> | ||
| + | <h3>Experiment</h3> | ||
| + | <br> | ||
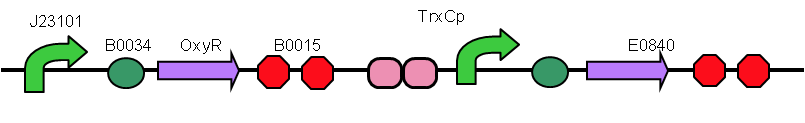
| + | <p>We sent our new sequence to Genscript China to be sequenced, along with the original OxyR sequence. The circuit we used to measure any improvement in our cells is found below and consists of a constitutive promoter which always produced OxyR transcription factor protein plus a TrxC promoter (part BBa_K1104201) which contains two binding sites for OxyR and is activated for expression of upstream genes once OxyR reacts with ROS and forms the intermolecular disulfide bond. Finally to be able to measure the effectiveness of this system to be induced by ROS, a reporter gene GFP is added and is under control of the inducible promoter TrxCp (part BBa_K1104201). This composite part, although was theoretically described by NYMU iGEM 2013, was constructed and out into the registry as a new composite part (BBa_K3031020) by us (SUIS-Shanghai iGEM 2019). We provide characterization of the composite part below which also provides useful descriptions for the parts BBa_K1104200 (OxyR coding sequence) and BBa_K1104201 (TrxC promoter)</p> | ||
| + | <figure class="figure-center"> | ||
| + | <img src="https://2019.igem.org/wiki/images/9/91/T--SUIS_Shanghai--_CompositeTrxCp.png" align="center" alt="H2O2 sensor chart"> | ||
| + | </figure> | ||
| + | <br> | ||
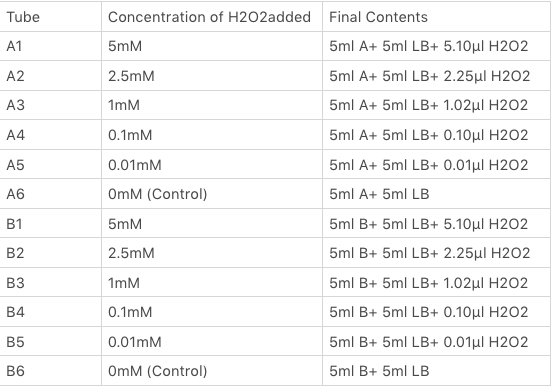
| + | <p>The OxyR gene is expressed by a constitutive promoter while the inducible promoter TrxC will only express downstream genes (GFP in this case) when it is activated by oxidized form of OxyR, by virture of binding to sites (maked in pink here). When H2O2 levels are high enough the OxyR is activated and GFP is produced. Both constructs were sequenced onto a expression vector pET301(+) and transformed the plasmid into Bl21(DE3) cells. To induce the TrxC promoter we divided four 50ml test tubes of OxyR and OxyR_Mutated into twelve 15ml test tubes, each contained 5ml culture broth and 5ml LB with OD600 0.4. H2O2 was added to 10 of the tubes in the following concentrations:</p> | ||
| + | <figure class="figure-center"> | ||
| + | <img src="https://2019.igem.org/wiki/images/1/15/T--SUIS_Shanghai--H2O2conc.png" height=300px width=400px alt="H2O2 sensor chart"> | ||
| + | </figure> | ||
| + | <p>Where:</p> | ||
| + | <ul> | ||
| + | <li><b>Sample A</b> = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing: Composite part with original gene sequence of OxyR (this part) under influence of constitutive promoter and GFP under the influence of inducible promoter TrxCp</li> | ||
| + | <li><b>Sample B<b> = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing: Composite part with mutated gene sequence of OxyR (BBa_K3031018) under influence of constitutive promoter and GFP under the influence of inducible promoter TrxCp</li> | ||
| + | <li><b>Blank<b> = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing no engineered plasmid. </li></ul> | ||
| + | <br> | ||
| + | <figure class="figure-center"> | ||
| + | <img src="https://2019.igem.org/wiki/images/8/8d/T--SUIS_Shanghai--H2O2assay.png" height=350px width=470px align="center" alt="H2O2 sensor chart"> | ||
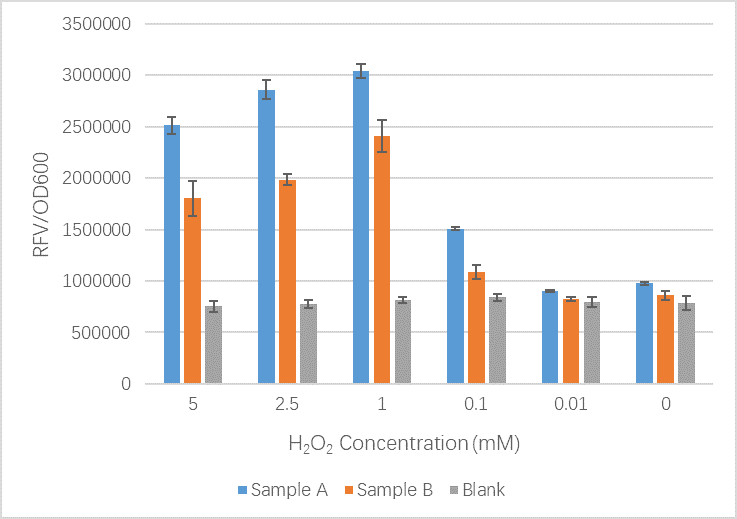
| + | <figcaption>Our results here show that our new part including mutations shows no significant difference between the original OxyR part at low H2O2 concentrations. When the concentration of H2O2 increases we see that the GFP signal is less than that of the original OxyR part. Our team believe that our new part is working well but we have realized that the slight conformation changes we likely introduced to the OxyR transcription factor protein may have meant it bind less tightly to the sites upstream of TrxC promoter and therefore expression is reduced. In this case we cannot be sure that our new part is more or less sensitive to oxidation by ROS.</figcaption> | ||
</figure> | </figure> | ||
<br> | <br> | ||
Latest revision as of 18:04, 21 October 2019
OxyR mutated at residues 147 & 203
OxyR is a Transcription factor protein. OxyR reacts with H2O2 resulting in the oxidation of a reactive cysteine (Cys-199) formation of a intermolecular disulfide bond with neighboring cyctein-208 and thus a conformation change in the OxyR protein occurs. This change is shape activates the OxyR protein which then can bind to certain promoters with binding site regions to regulate the expression of downstream genes.

Biology & Uses
This part sequence is a modified version of the OxyR gene sequence (BBa_K1104200) which was submitted by National Yang Ming University iGEM Team in 2013. We attempted to improve the sensitivity of this OxyR to reactive oxygen species (ROS) by editing the sequence to change targeted amino acids surrounding the cysteine-199, thus increasing the accessible surface area of the reactive cysteine (cysteine-199) within the protein and making it more susceptible to oxidation. We hoped that our mutated protein would show a higher sensitivity to low concentration of H2O2, which is a sign of cellular stress and perhaps an immune response to infection from a pathogen. We hoped to make this transcription factor more sensitive to lower concentrations of H2O2 and potentially create a new device which could be used to be a very sensitive indicator of early immune response and infection.
Part Design
Our approach involved using site specific structural data gathered from high resolution crystal images of proteins shown to have an oxidized cysteine (i.e. formed a sulfenic acid). The environments of these resides were explored and analyzed and compared to non-oxidized cysteines within the same proteins. Differences between the environments of oxidized and reduced cysteines that were discovered between these two groups were taken as indicators of facilitating oxidation and therefore could potentially be used to modify OxyR protein and make it more sensitive to oxidative stress. High-resolution crystal structures of proteins (2.2 Angstroms resolution or better) containing S-sulfenylated cysteine sites were first obtained from the PDB (www.pdb.org) and multiple sequence alignment was used to ensure that only unique protein microenvironments would be analyzed. A representative list of unmodified cysteine sites was identified for comparative analysis with the modified sites by first profiling the proteins containing S-sulfenylated sites by UnipriotKB GO annotations (https://www.uniprot.org/uploadlists/) and then by choosing 40% of proteins from the original S-sulfenylated cysteine protein list which contained unmodified cysteines. In total the number of unique S-sulfeylated cysteine environments analyzed was 373 from 322 proteins. While using the selection criteria outlined in above, a total of 426 unmodified cysteine sites were analyzed in comparison.

The Amino acid, solvent, and ligand contacts for each atom contained within all Cys-SOH and unmodified cysteine sites in the dataset was furthered analyzed using PISA (Protein Interphases, Surfaces and Assemblies) (Krissinel & Henrick, 2007). The CCP4 Graphical User Interface (CCP4i) (Potterton, Briggs, Turkenburg, & Dodson, 2003) was used to perform and visualize PISA calculations. Crystal structures of protein containing sites of interest in PDB format were used as inputs into the interface. Default parameters were used and only calculations for contacts between 2.0 to 3.2 Angstroms within the chain containing the residue of interest were obtained.


Amino Acid composition surrounding the 3.5A space around the S-sulfenylated cystienes (left) and th unmodified cysteines (right)
The solvent accessible surface area (SASA) of Cys-199 was determined using AREAMOL. Areaimol is a tool from the collection of programs from the Collaborative Computational Project No. 4 (CCP4) software suite version 7.0 (Winn et al., 2011). It finds the solvent accessible area of atoms in a PDB coordinate file by calculating the area of an atoms Van der Waals surface that a probe sphere rolls over (Lee & Richards, 1971). The default probe radius of 1.4 Angstroms was used to determine SASA. The CCP4 Graphical User Interface (CCP4i) (Potterton et al., 2003) was used to perform AREAIMOL calculations.
SASA of cysteine-199 in reduced form (PDB ID: 1i69) = 21.5 Angstroms
Relative solvent accessibility (RSA) was determined by dividing the SASA by the max accessible surface area (maxASA).
- RSA = SASA/maxASA
- where: RSA = relative Solvent Accessibility
- SASA = Solvent Accessibly Surface Area
- maxASA = maximum Accessible Surface Area
The maxASA value used for cysteine 167.0 Angstroms (Tian et al., 2013). An RSA of 10% was used to be the cut off for classifying a residue as surface accessible (RSA > 10%) or buried (RSA < 10%). A comparison of modified and unmodified cysteine RSA values was conducted in order to determine if there was enrichment for exposed or buried surfaces within the Cys-SOH environments. A quantile– quantile probability plot (Q-Q plot) was used to determine if there was enrichment for exposed or buried cysteines within the Cys-SOH compared to unmodified sites. RSA values of modified and unmodified cysteine residues were sorted and values of RSA for each quantile was plotted against each other (i.e. RSA values of modified and unmodified cysteine residues were sorted from 0.01 to 1.0 and plotted against each other).

Mutations made to OxyR (part BBa_K1104200):
- Residue 203: Glutamine (wild type) mutated to Serine Reasoning: Serine is overrepresented at position + 4 in the amino acid sequences flanking the S-sulfenylated cysteines (fig 7). In OxyR this position was filled with a Glutamine residue. We decided that placing a Serine there may allow for an improvement sensitivity to oxidation. From the literature accompanied by our modeling it was found that Serine, as a residue being an uncharged H-bond donor, likely has an important role in the activation of Cysteine, mainly by lowering its pKa (Mariano & Gladyshev, 2011). These two pieces of evidence allowed us to make this decision to mutate the protein.
- Residue 149: Valine (wild type) mutated to Glycine We replaced on hydrophobic residue with another smaller sized one in the hope of increasing the solvent accessibility on the cysteine’s thiol. Val-149 was in close contact with Cys-199 as described below.
Experiment
We sent our new sequence to Genscript China to be sequenced, along with the original OxyR sequence. The circuit we used to measure any improvement in our cells is found below and consists of a constitutive promoter which always produced OxyR transcription factor protein plus a TrxC promoter (part BBa_K1104201) which contains two binding sites for OxyR and is activated for expression of upstream genes once OxyR reacts with ROS and forms the intermolecular disulfide bond. Finally to be able to measure the effectiveness of this system to be induced by ROS, a reporter gene GFP is added and is under control of the inducible promoter TrxCp (part BBa_K1104201). This composite part, although was theoretically described by NYMU iGEM 2013, was constructed and out into the registry as a new composite part (BBa_K3031020) by us (SUIS-Shanghai iGEM 2019). We provide characterization of the composite part below which also provides useful descriptions for the parts BBa_K1104200 (OxyR coding sequence) and BBa_K1104201 (TrxC promoter)
The OxyR gene is expressed by a constitutive promoter while the inducible promoter TrxC will only express downstream genes (GFP in this case) when it is activated by oxidized form of OxyR, by virture of binding to sites (maked in pink here). When H2O2 levels are high enough the OxyR is activated and GFP is produced. Both constructs were sequenced onto a expression vector pET301(+) and transformed the plasmid into Bl21(DE3) cells. To induce the TrxC promoter we divided four 50ml test tubes of OxyR and OxyR_Mutated into twelve 15ml test tubes, each contained 5ml culture broth and 5ml LB with OD600 0.4. H2O2 was added to 10 of the tubes in the following concentrations:
Where:
- Sample A = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing: Composite part with original gene sequence of OxyR (this part) under influence of constitutive promoter and GFP under the influence of inducible promoter TrxCp
- Sample B = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing: Composite part with mutated gene sequence of OxyR (BBa_K3031018) under influence of constitutive promoter and GFP under the influence of inducible promoter TrxCp
- Blank = E.coli BL21(DE3) cells containing expression plasmid pET30a(+) containing no engineered plasmid.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 720
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 673
