Difference between revisions of "Part:BBa K1150000"
| Line 54: | Line 54: | ||
https://2019.igem.org/wiki/images/6/6a/T--Peking--picture_for_ssrA_new.jpeg | https://2019.igem.org/wiki/images/6/6a/T--Peking--picture_for_ssrA_new.jpeg | ||
| − | Figure 1: Characterization of CRISPRri-ssrA system. | + | Figure 1: Characterization of CRISPRri-ssrA system. (A) Comparison between dCas9 and dCas9-ssrA system by expression level and CRISPRi effect on mRFP fluorescence. (B) Comparison of effect on cell growth between CRISPRri and CRISPRri-ssrA, both targeted to R1+ box(left:R1+; right:R1+ ssrA). (C) Reversibility of CRISPRri-ssrA system targeted to R1+ box. Hollow arrows stand for removal of arabinose while solid black arrows stand for addition of arabinose. |
| − | A | + | |
| − | B | + | |
| − | C | + | |
Reference: | Reference: | ||
Revision as of 17:30, 21 October 2019
dCas9
| dCas9 | |
|---|---|
| Function | Binding protein |
| Use in | Mammalian cells |
| RFC standard | RFC 25 |
| Backbone | pSB1C3 |
| Organism | Streptococcus pyogenes |
| Source | Feng Zhang, Addgene |
| Submitted by | [http://2013.igem.org/Team:Freiburg Freiburg 2013] |
dCas9 is a codon optimized and standardized (RFC 25) protein for human cell lines. Interacting with a DNA-binding RNA and fused with different effector domains it can be used for specific gene regulation.
Cas9 is the main protein of the CRISPR/Cas system II of Streptococcus pyogenes. CRISPR systems protect bacteria and archaea from phages by recognizing and cleaving of invading phage DNA. This recognition is based on Watson Crick base pairing between a short RNA, called crRNA, and the complementary DNA strand. A second RNA, called tracrRNA, connects crRNA and Cas9. These three parts together form a protein-RNA-DNA complex with the targeted DNA strand [1].
Cas9 became of great interest for research concerning DNA targeting, because of its ability to recognize site specific DNA strands by a crRNA.
At first the functionality of Cas9 was modified by exchanging aminoacids. As a result, Cas9 was able to introduce mutations within the genome of several organisms by causing double strand breaks [2][3]. Then, it was converted from a nuclease to a nickase introducing single strand breaks [4] and lately it was converted to an enzymatically inactive form, called dCas9 [5].
This dCas9 is codon optimized for human cell lines and standardized (RFC 25). It can be used as a DNA binding protein, that can be fused with different effectors in order to regulate gene expression.
Improvement
Characterized by Peking 2019
In our experiment, we found that the inhibitory effect of dCas9 to DNA replication shows to have over-inhibition on cell growth and small dynamic range when the sgRNAs targeting high affinity DnaA boxes. In order to extend this system to combine all DnaA boxes, we improve the performance of the CRISPRri system by weakening its effect by adding a degradation signal peptide ssrA to dCas9. We get an improved part (BBa_K3081055) from dCas9 (BBa_K1150000). The ssrA peptide tag from Mycoplasma florum has been developed as a versatile biotechnology tool to control orthogonal degradation of tagged proteins in Escherichia coli [1]. Ideally, after fused dCas9 and ssrA, it accelerates the degradation rate of dCas9 and thus weaken the inhibition of DNA replication by CRISPRri. As a preliminary verification, firstly, we fused ssrA tag to dCas9-sfGFP, we tested the expression curve of dCas9-sfGFP-ssrA by adding different concentration of inducer (Figure 1A). We found that the protein with ssrA was expressed at a lower level than the control protein, with the same concentration of inducer. We believe that because ssrA mediates protein degradation, the equilibrium concentration of the protein is reduced. To test the function of fused version of dCas9-ssrA, we co-transformed two plasmids to one E. coli cell, which express mRFP and dCas9-ssrA with sgRNA targeting CDS of mRFP, respectively. As a matter of fact, the new CRISPRi system shows a gentler decrease in fluorescence when dCas9 is fused with ssrA tag, while non-binding dCas9 with or without ssrA has no influence on mRFP expression (Figure 1B). This result confirmed fusion with ssrA, the original function of dCas9 is unaffected. So, we tested the improved CRISPRri-ssrA system with target site to DnaA boxes which are shown to have excessive inhibition on cell growth before. We found that the degradation tag make inhibition effect much milder, which allows for a wider adjusting range (Figure 1C).
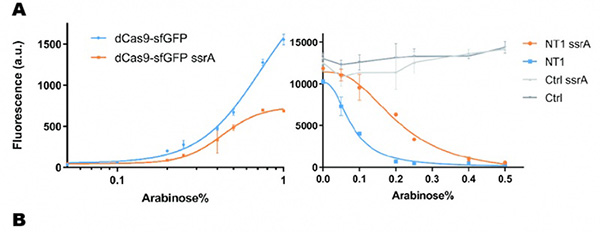


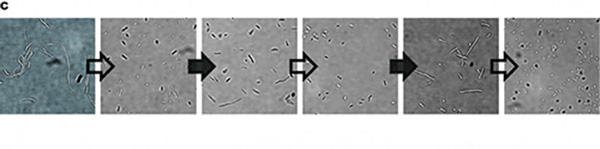
Figure 1: Characterization of CRISPRri-ssrA system. (A) Comparison between dCas9 and dCas9-ssrA system by expression level and CRISPRi effect on mRFP fluorescence. (B) Comparison of effect on cell growth between CRISPRri and CRISPRri-ssrA, both targeted to R1+ box(left:R1+; right:R1+ ssrA). (C) Reversibility of CRISPRri-ssrA system targeted to R1+ box. Hollow arrows stand for removal of arabinose while solid black arrows stand for addition of arabinose.
Reference:
[1] Lv, L., Wu, Y., Zhao, G., & Qi, H. (2019). Improvement in the Orthogonal Protein Degradation in Escherichia coli by Truncated mf-ssrA Tag. Transactions of Tianjin University, 1-7.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 248
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
| Protein data table for BioBrick BBa_K1150000 automatically created by the BioBrick-AutoAnnotator version 1.0 | ||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide sequence in RFC 25, so ATGGCCGGC and ACCGGT were added (in italics) to the 5' and 3' ends: (underlined part encodes the protein) ATGGCCGGCGACAAGAAG ... GGAGGCGACACCGGT ORF from nucleotide position -8 to 4107 (excluding stop-codon) | ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid sequence: (RFC 25 scars in shown in bold, other sequence features underlined; both given below)
| ||||||||||||||||||||||||||||||||||||||||||||||
Sequence features: (with their position in the amino acid sequence, see the list of supported features)
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid composition:
| ||||||||||||||||||||||||||||||||||||||||||||||
Amino acid counting
| Biochemical parameters
| |||||||||||||||||||||||||||||||||||||||||||||
| Plot for hydrophobicity, charge, predicted secondary structure, solvent accessability, transmembrane helices and disulfid bridges | ||||||||||||||||||||||||||||||||||||||||||||||
Codon usage
| ||||||||||||||||||||||||||||||||||||||||||||||
| Alignments (obtained from PredictProtein.org) There were no alignments for this protein in the data base. The BLAST search was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| Predictions (obtained from PredictProtein.org) | ||||||||||||||||||||||||||||||||||||||||||||||
| There were no predictions for this protein in the data base. The prediction was initialized and should be ready in a few hours. | ||||||||||||||||||||||||||||||||||||||||||||||
| The BioBrick-AutoAnnotator was created by TU-Munich 2013 iGEM team. For more information please see the documentation. If you have any questions, comments or suggestions, please leave us a comment. | ||||||||||||||||||||||||||||||||||||||||||||||
References
[1] Westra E.R., Swarts D.C., Staals R.H., Jore M.M., Brouns S.J., van der Oost J. (2012). The CRISPRs, they are a-changin': how prokaryotes generate adaptive immunity. Annu Rev Genet. 46, 311-39
[2] Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. (2013). RNA-guided human genome engineering via Cas9. Science 339(6121), 823-6
[3] Jiang W., Bikard D., Cox D., Zhang F., Marraffini L.A. (2013). RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat Biotechnol. 31(3), 233-9
[4] Cong, L., Ran, F.A., Cox, D., Lin, S., Barretto, R., Habib, N., Hsu, P.D., Wu, X., Jiang, W., Marraffini, L.A., Zhang, F. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339 (6121), 819-23
[5] Qi L.S., Larson M.H., Gilbert L.A., Doudna J.A., Weissman J.S., Arkin A.P., Lim W.A. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152(5), 1173-83
