Difference between revisions of "Part:BBa K3282003"
| (6 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3282003 short</partinfo> | <partinfo>BBa_K3282003 short</partinfo> | ||
| − | The part consists of T7 promoter (BBa_I719005), Anderson RBS (BBa_J61109), arsenic resistance operon | + | |
| + | The part consists of T7 promoter [https://parts.igem.org/Part:BBa_I719005 (Part:BBa_I719005)], Anderson RBS [https://parts.igem.org/Part:BBa_J61109 (Part:BBa_J61109)], arsenic resistance operon repressor [https://parts.igem.org/Part:BBa_K3282000 (Part:BBa_K3282000)] and TE terminator [https://parts.igem.org/Part:BBa_B0012 (Part:BBa_B0012)]. | ||
| + | The ''Escherichia coli'' chromosomal ''ars'' operon confers resistance to arsenicals by a specific efflux pump controlled by an arsenite-inducible repressor, ''arsR'' which has been shown to be a trans-acting repressor that senses environmental As(III)[1]. | ||
| + | |||
| + | '''Cultivation of Transformed Bacteria''' | ||
| + | |||
| + | The E. coli BL21(DE3) strain was transformed with biobrick ars-T7 in pUC19 plasmid by heat shock. The growth rate of the transformed and wild type strains were compared by cultivating the cells in LB media at 37 °C and measuring OD every hour. The cultivation was carried out to investigate if the presence of plasmid acts as a burden for the cells. As is evident from '''Figure 1''', the growth rate of the control and the transformed bacteria are very similar. This is unexpected since ''arsR'' expression has shown to be toxic to cells [4]. This conclusion is a positive indicator that the cells would not die before their intended use of absorbing arsenic. | ||
| + | |||
| + | [[File:ArsT7cultivation.png]] | ||
| + | |||
| + | '''Figure 1: Comparison of the growth rate of E. coli BL21(DE3) transformed with ars-T7 inserted into pUC19 and wild type E. coli BL21(DE3).''' | ||
| + | |||
| + | '''Toxic Metal Analysis''' | ||
| + | |||
| + | This part was ligated into pUC19 and transformed into ''E. coli'' BL21(DE3). The expression of ''arsR'' was demonstrated by performing Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) analysis. The cells were cultured in enriched media (containing peptone, yeast extract, phosphates, chlorides and sulphates) at 37 °C. They were grown overnight followed by cell harvesting to normalize the OD values to 2. After normalization, the cells were resuspended in fresh enriched medium containing 10 µM, 50 µM and 200 µM of As2O3. As a negative control, the experiment was also performed with the non engineered ''E. coli'' BL21(DE3) in growth medium containing 50 µM As2O3. | ||
| + | Samples were collected every hour for OD measurement. For investigating the arsenic accumulation by engineered bacteria, samples were collected at 0 hour, 2.5 hour and 5 hour after the addition of As2O3. | ||
| + | |||
| + | |||
| + | '''Results''' | ||
| + | |||
| + | The SDS-PAGE analysis result can be seen in '''Figure 2''' . ''arsR'' has a molecular weight of 13.5 kDa, which is expected to be between the two bands marked in '''Figure 2'''. The gel shows that the engineered bacteria as well as the negative control have a band with the expected molecular weight. Thus, we cannot confirm that it corresponds to ''arsR''. It is possible that there are other proteins expressed in ''E. coli'' of the same molecular weight, masking the expression of ''arsR''. | ||
| + | |||
| + | |||
| + | [[File:K3282003-SDS.png]] | ||
| + | |||
| + | '''Figure 2: Samples were harvested at different time points, and the total cellular proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein bands of BL21(DE3) can be seen after induction with IPTG, showing ''arsR'' at 13.5 kDa. The control shown is non-engineered BL21(DE3) without ''arsR''.''' | ||
| + | |||
| + | |||
| + | [[File:K3282003-TM.png]] | ||
| + | |||
| + | '''Figure 3: Comparison of arsenite concentration in the growth medium containing ''E. coli'' BL21(DE3) cells expressing ''arsR'' and ''E. coli'' BL21(DE3) cells without arsR expression.''' | ||
| + | |||
| + | |||
| + | [[File:K3282003-OD.png]] | ||
| + | |||
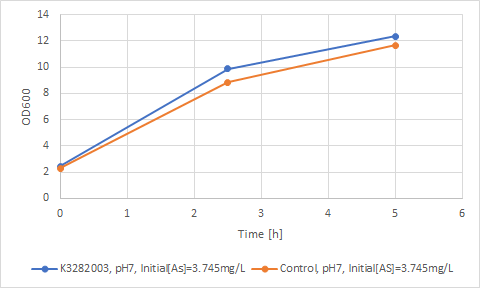
| + | '''Figure 4: Comparison of growth rate of engineered BL21(DE3) with wild type BL21(DE3).''' | ||
| + | |||
| + | |||
| + | The '''Figure 3''' compares the arsenite accumulation over time whereas '''Figure 4''' compares the growth rate of engineered as well as non-engineered BL21(DE3). It can be seen that the initial added arsenite had the concentration of 3.745 mg/L for both engineered as well as non-engineered BL21(DE3) and the arsenite concentration measured at 0 hour was 0.014 mg/L and 0.013 mg/L respectively. This rapid decrease in the concentration was not expected. This rapid decrease cannot be accounted for by the sticking of ions to the glass surface since it has been shown that arsenite does not suffer from sorption to the glass surfaces as it forms oxy ions which is partly dissociated leading to negatively charged ions [2].It might be the case that the engineered bacteria as well as the control have a capacity to uptake arsenite. According to Chen et al., when the cells are depleted of endogenous energy reserves, arsenite enters whether or not the cells carry a resistance plasmid [3]. The ''ars'' operon carried on the ''Escherichia coli'' R factor R773 encodes the transport system that extrudes arsenite and the lowering of the intracellular concentration of toxic oxyanion produces resistance, however, plasmidless strains of ''E. coli'' have also been shown to be intrinsically resistant to arsenite due to the presence of chromosomal ''ars'' operon. It has also been found that the R773 ''arsR'' protein is 75% identical with the chromosomal product in ''E. coli'' strains [4]. The wild type ''E. coli'' strains are resistant to arsenite upto the concentration of 1 mM. This might explain the similarity in growth and arsenite uptake between the engineered strain and the wild type strain. However, if the measured arsenite concentration at 0 hour is compared with the final concentration at 5 hour, the engineered BL21(DE3) shows a 21.43 % arsenite accumulation compared with only 7.69 % arsenite accumulation in the control. This can be considered as a proof of concept showing that the engineered bacteria express ''arsR'' and has an improved ability to uptake arsenite. | ||
| + | |||
| + | |||
| + | '''Future Improvements''' | ||
| + | |||
| + | Since there was no visible protein expression at the expected position, one could try to add a histidine tag, making purification of protein possible and one would be able to assess the protein expression. Another solution would be to use a stronger promoter, which would give a higher expression of protein, and might be visible on the SDS-PAGE. Another improvement could be to make sure the protein is not stuck in the wells of the gel. According to Kostal et al. (2004) overexpression of ''arsR'' can be toxic to the host. It was found that fusing ''arsR'' with elastin polypeptide or maltose binding protein can reduce the toxic effect and hence improve the overexpression of arsenic accumulation protein. | ||
| + | |||
| + | |||
| + | '''References''' | ||
| + | |||
| + | 1. Kostal, J., Yang, R., Wu, C. H., Mulchandani, A., & Chen, W. (2004). Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR. Applied and environmental microbiology, 70(8), 4582–4587. doi:10.1128/AEM.70.8.4582-4587.2004. | ||
| + | |||
| + | 2. Massee, R., Maessen, F. (1981). Losses of silver, arsenic, cadmium, selenium, and zinc traces from distilled water and artificial sea-water by sorption on various container surface. Analytica Chimica Acta, 127, pp. 206-210. | ||
| + | |||
| + | 3. Chen, C., Misra, T., Silver, S., Rosen, B. Nucleotide sequence of the structural genes for an anion pump: the plasmid-encoded arsenical resistance operon. (1986). J. Biol. Chem., 261, pp. 15030-15038. | ||
| + | |||
| + | 4. Carlin, A., Shi, W., Dey, S., & Rosen, B. P. (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. Journal of bacteriology, 177(4), 981–986. doi:10.1128/jb.177.4.981-986.1995. | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 16:16, 21 October 2019
ArsR under T7 promoter
The part consists of T7 promoter (Part:BBa_I719005), Anderson RBS (Part:BBa_J61109), arsenic resistance operon repressor (Part:BBa_K3282000) and TE terminator (Part:BBa_B0012).
The Escherichia coli chromosomal ars operon confers resistance to arsenicals by a specific efflux pump controlled by an arsenite-inducible repressor, arsR which has been shown to be a trans-acting repressor that senses environmental As(III)[1].
Cultivation of Transformed Bacteria
The E. coli BL21(DE3) strain was transformed with biobrick ars-T7 in pUC19 plasmid by heat shock. The growth rate of the transformed and wild type strains were compared by cultivating the cells in LB media at 37 °C and measuring OD every hour. The cultivation was carried out to investigate if the presence of plasmid acts as a burden for the cells. As is evident from Figure 1, the growth rate of the control and the transformed bacteria are very similar. This is unexpected since arsR expression has shown to be toxic to cells [4]. This conclusion is a positive indicator that the cells would not die before their intended use of absorbing arsenic.
Figure 1: Comparison of the growth rate of E. coli BL21(DE3) transformed with ars-T7 inserted into pUC19 and wild type E. coli BL21(DE3).
Toxic Metal Analysis
This part was ligated into pUC19 and transformed into E. coli BL21(DE3). The expression of arsR was demonstrated by performing Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) analysis. The cells were cultured in enriched media (containing peptone, yeast extract, phosphates, chlorides and sulphates) at 37 °C. They were grown overnight followed by cell harvesting to normalize the OD values to 2. After normalization, the cells were resuspended in fresh enriched medium containing 10 µM, 50 µM and 200 µM of As2O3. As a negative control, the experiment was also performed with the non engineered E. coli BL21(DE3) in growth medium containing 50 µM As2O3. Samples were collected every hour for OD measurement. For investigating the arsenic accumulation by engineered bacteria, samples were collected at 0 hour, 2.5 hour and 5 hour after the addition of As2O3.
Results
The SDS-PAGE analysis result can be seen in Figure 2 . arsR has a molecular weight of 13.5 kDa, which is expected to be between the two bands marked in Figure 2. The gel shows that the engineered bacteria as well as the negative control have a band with the expected molecular weight. Thus, we cannot confirm that it corresponds to arsR. It is possible that there are other proteins expressed in E. coli of the same molecular weight, masking the expression of arsR.
Figure 2: Samples were harvested at different time points, and the total cellular proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Protein bands of BL21(DE3) can be seen after induction with IPTG, showing arsR at 13.5 kDa. The control shown is non-engineered BL21(DE3) without arsR.
Figure 3: Comparison of arsenite concentration in the growth medium containing E. coli BL21(DE3) cells expressing arsR and E. coli BL21(DE3) cells without arsR expression.
Figure 4: Comparison of growth rate of engineered BL21(DE3) with wild type BL21(DE3).
The Figure 3 compares the arsenite accumulation over time whereas Figure 4 compares the growth rate of engineered as well as non-engineered BL21(DE3). It can be seen that the initial added arsenite had the concentration of 3.745 mg/L for both engineered as well as non-engineered BL21(DE3) and the arsenite concentration measured at 0 hour was 0.014 mg/L and 0.013 mg/L respectively. This rapid decrease in the concentration was not expected. This rapid decrease cannot be accounted for by the sticking of ions to the glass surface since it has been shown that arsenite does not suffer from sorption to the glass surfaces as it forms oxy ions which is partly dissociated leading to negatively charged ions [2].It might be the case that the engineered bacteria as well as the control have a capacity to uptake arsenite. According to Chen et al., when the cells are depleted of endogenous energy reserves, arsenite enters whether or not the cells carry a resistance plasmid [3]. The ars operon carried on the Escherichia coli R factor R773 encodes the transport system that extrudes arsenite and the lowering of the intracellular concentration of toxic oxyanion produces resistance, however, plasmidless strains of E. coli have also been shown to be intrinsically resistant to arsenite due to the presence of chromosomal ars operon. It has also been found that the R773 arsR protein is 75% identical with the chromosomal product in E. coli strains [4]. The wild type E. coli strains are resistant to arsenite upto the concentration of 1 mM. This might explain the similarity in growth and arsenite uptake between the engineered strain and the wild type strain. However, if the measured arsenite concentration at 0 hour is compared with the final concentration at 5 hour, the engineered BL21(DE3) shows a 21.43 % arsenite accumulation compared with only 7.69 % arsenite accumulation in the control. This can be considered as a proof of concept showing that the engineered bacteria express arsR and has an improved ability to uptake arsenite.
Future Improvements
Since there was no visible protein expression at the expected position, one could try to add a histidine tag, making purification of protein possible and one would be able to assess the protein expression. Another solution would be to use a stronger promoter, which would give a higher expression of protein, and might be visible on the SDS-PAGE. Another improvement could be to make sure the protein is not stuck in the wells of the gel. According to Kostal et al. (2004) overexpression of arsR can be toxic to the host. It was found that fusing arsR with elastin polypeptide or maltose binding protein can reduce the toxic effect and hence improve the overexpression of arsenic accumulation protein.
References
1. Kostal, J., Yang, R., Wu, C. H., Mulchandani, A., & Chen, W. (2004). Enhanced arsenic accumulation in engineered bacterial cells expressing ArsR. Applied and environmental microbiology, 70(8), 4582–4587. doi:10.1128/AEM.70.8.4582-4587.2004.
2. Massee, R., Maessen, F. (1981). Losses of silver, arsenic, cadmium, selenium, and zinc traces from distilled water and artificial sea-water by sorption on various container surface. Analytica Chimica Acta, 127, pp. 206-210.
3. Chen, C., Misra, T., Silver, S., Rosen, B. Nucleotide sequence of the structural genes for an anion pump: the plasmid-encoded arsenical resistance operon. (1986). J. Biol. Chem., 261, pp. 15030-15038.
4. Carlin, A., Shi, W., Dey, S., & Rosen, B. P. (1995). The ars operon of Escherichia coli confers arsenical and antimonial resistance. Journal of bacteriology, 177(4), 981–986. doi:10.1128/jb.177.4.981-986.1995.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 192
- 1000COMPATIBLE WITH RFC[1000]