Difference between revisions of "Part:BBa K3221009"
LiuTianAlice (Talk | contribs) |
|||
| (13 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K3221009 short</partinfo> | <partinfo>BBa_K3221009 short</partinfo> | ||
| − | We constructed this part on pYES2 to express HisG-MF in the yeast | + | We constructed this part on pYES2 to express HisG-MF in the yeast BY4741. |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 17: | Line 17: | ||
<partinfo>BBa_K3221009 parameters</partinfo> | <partinfo>BBa_K3221009 parameters</partinfo> | ||
<!-- --> | <!-- --> | ||
| − | + | <div style="font-size:17px;"> | |
| − | '''iGEM19_SCU-China:''' | + | '''iGEM19_SCU-China:''' |
| − | < | + | </div> |
| + | <div style="font-size:15px;"> | ||
This part is pGAL1-HisG-MF, which is a CDS of HisG-MF with a pGAL1 promoter upstream and a CYC1 terminator downstream. HisG-MF is half of Cns3 containing the HisG domain, which is designed from the reference and has been validated by 2019 SCU-China for expression and function in other fungus. | This part is pGAL1-HisG-MF, which is a CDS of HisG-MF with a pGAL1 promoter upstream and a CYC1 terminator downstream. HisG-MF is half of Cns3 containing the HisG domain, which is designed from the reference and has been validated by 2019 SCU-China for expression and function in other fungus. | ||
Cns3 is an enzyme originated from ''Cordyceps militaris'', it catalyzes the biosynthesis of Pentostatin (2’-deoxycoformycin, PTN), an ada chemotherapeutic drug used for the treatment of several forms of leukemia.It has been proved that PTN can protect the stability of cordycepin from deamination by inhibiting adenosine deaminase (ADA) activity in the natural pathway of COR in ''C.militaris''.Therefore, in our designs, we make use of Cns3, producing PTN together with COR in order to guarantee a high yield of COR.Previous studies have also proved that the NK domain of Cns3, one of it's two function domains, is responsible for PTN biosynthesis, which is why we only use part of Cns3. | Cns3 is an enzyme originated from ''Cordyceps militaris'', it catalyzes the biosynthesis of Pentostatin (2’-deoxycoformycin, PTN), an ada chemotherapeutic drug used for the treatment of several forms of leukemia.It has been proved that PTN can protect the stability of cordycepin from deamination by inhibiting adenosine deaminase (ADA) activity in the natural pathway of COR in ''C.militaris''.Therefore, in our designs, we make use of Cns3, producing PTN together with COR in order to guarantee a high yield of COR.Previous studies have also proved that the NK domain of Cns3, one of it's two function domains, is responsible for PTN biosynthesis, which is why we only use part of Cns3. | ||
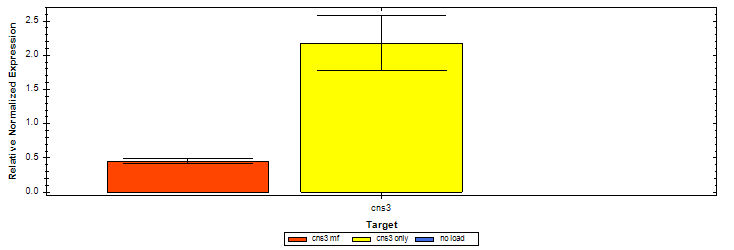
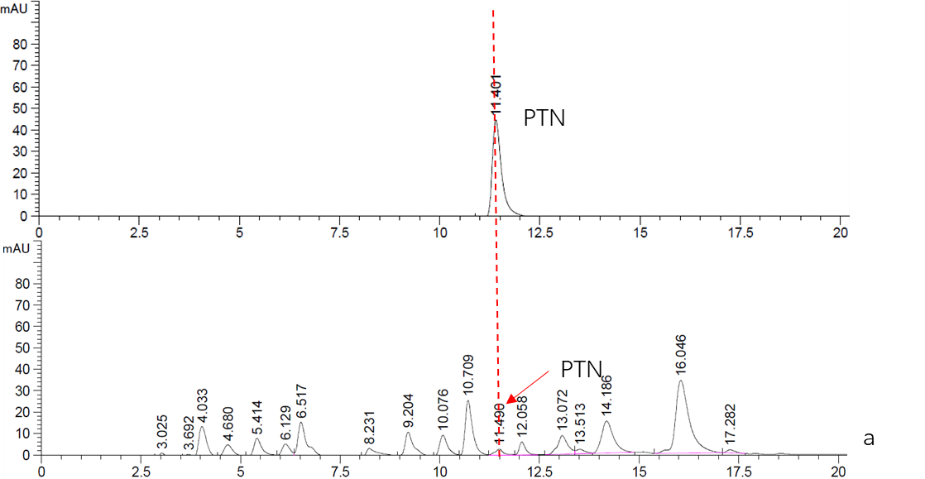
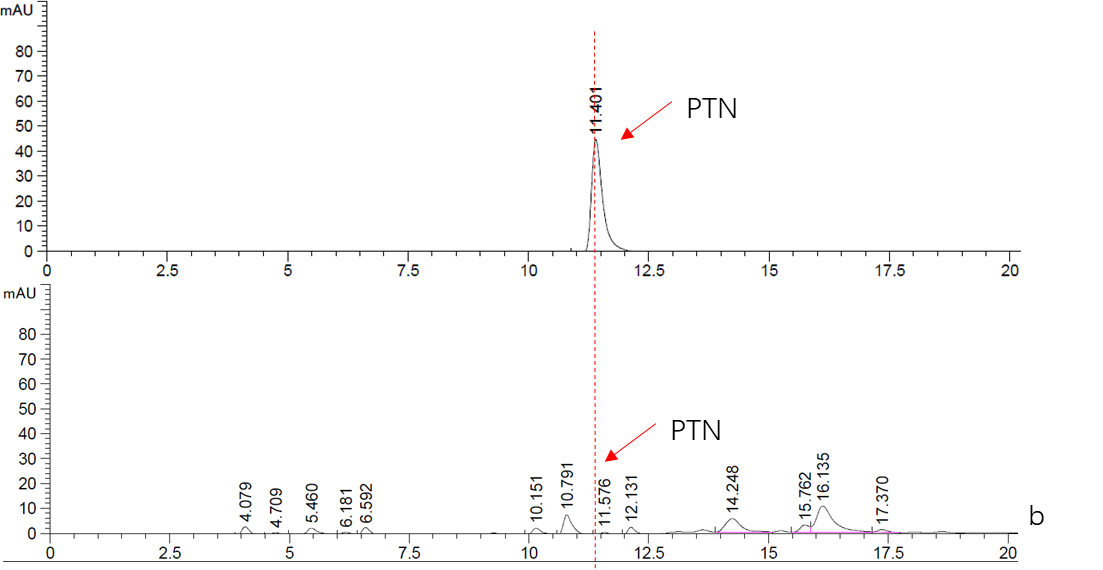
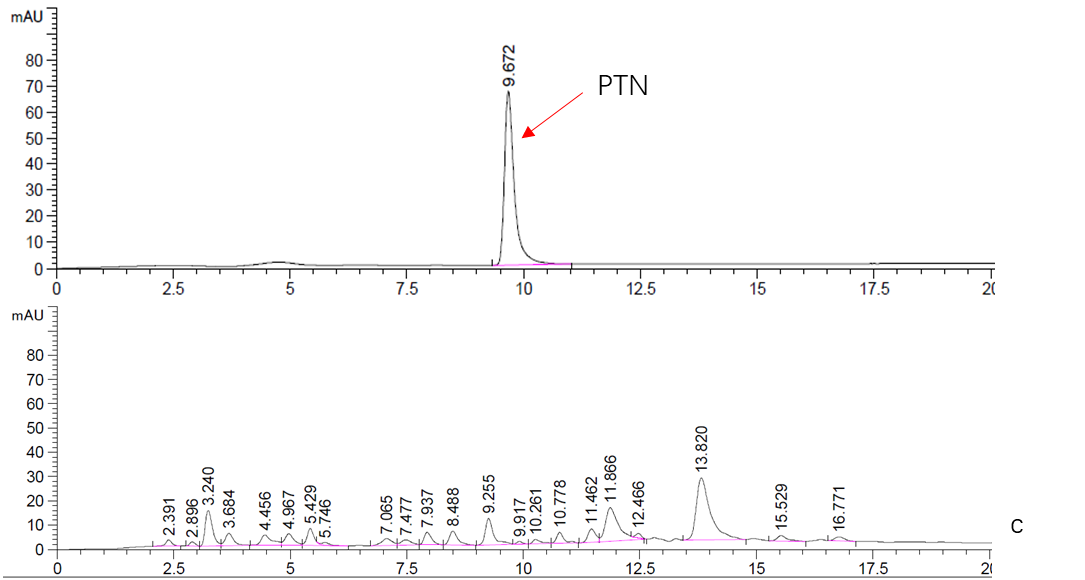
<p>As is mentioned above, this part is an expression cassette of HisG-MF that is half of Cns3 containing the HisG domain. 2019 SCU-China validated its both in expression and function in ''Saccharomyces cerevisiae''.To verify it’s function, we did RT-qPCR to detect the existence of Cns3-MF mRNA. (Figure. 1) As for function validation, we performed HPLC to detect PTN, the catalysate of Cns3-MF(Figure. 2). And we successfully demonstrated the cns3-mf transcription and function finally. </p> | <p>As is mentioned above, this part is an expression cassette of HisG-MF that is half of Cns3 containing the HisG domain. 2019 SCU-China validated its both in expression and function in ''Saccharomyces cerevisiae''.To verify it’s function, we did RT-qPCR to detect the existence of Cns3-MF mRNA. (Figure. 1) As for function validation, we performed HPLC to detect PTN, the catalysate of Cns3-MF(Figure. 2). And we successfully demonstrated the cns3-mf transcription and function finally. </p> | ||
| + | </div> | ||
| + | [[File:T--SCU-China--qPCR-Cns3.png|720px]] | ||
| − | + | <b>Figure. 1</b> '''RT-qPCR of Cns3-HisG-mf\Cns3-HisG-only.''' | |
| − | + | <br>No-load is the S. cerevisiae without transformed plasmids. The reference gene is PGK1. The RNA of cns3 is detected successfully. The bars are SE. | |
| − | <b>Figure. 1</b> '''RT-qPCR of | + | |
| − | <br>No-load is the S. cerevisiae without transformed plasmids. The reference gene is PGK1. The RNA of | + | |
[[File:T--SCU-China--scu-demonstration-fig4-2.png|720px]] | [[File:T--SCU-China--scu-demonstration-fig4-2.png|720px]] | ||
| Line 37: | Line 38: | ||
'''Figure. 2''' '''HPLC analysis of PTN''' | '''Figure. 2''' '''HPLC analysis of PTN''' | ||
| − | <br>a. Top: Standard sample of 5mM PTN. | + | <br>a. Top: Standard sample of 5mM PTN. |
<br>Bottom: Supernatant fraction of Cns3-hisG-MF sample. The strain BY4741 was culturing in YPDG media for 3 days. | <br>Bottom: Supernatant fraction of Cns3-hisG-MF sample. The strain BY4741 was culturing in YPDG media for 3 days. | ||
<br>b. Top: Standard sample of 5mM PTN. | <br>b. Top: Standard sample of 5mM PTN. | ||
Latest revision as of 15:36, 21 October 2019
pGAL1-HisG MF-tCYC1
We constructed this part on pYES2 to express HisG-MF in the yeast BY4741.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 682
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 150
- 1000COMPATIBLE WITH RFC[1000]
iGEM19_SCU-China:
This part is pGAL1-HisG-MF, which is a CDS of HisG-MF with a pGAL1 promoter upstream and a CYC1 terminator downstream. HisG-MF is half of Cns3 containing the HisG domain, which is designed from the reference and has been validated by 2019 SCU-China for expression and function in other fungus.
Cns3 is an enzyme originated from Cordyceps militaris, it catalyzes the biosynthesis of Pentostatin (2’-deoxycoformycin, PTN), an ada chemotherapeutic drug used for the treatment of several forms of leukemia.It has been proved that PTN can protect the stability of cordycepin from deamination by inhibiting adenosine deaminase (ADA) activity in the natural pathway of COR in C.militaris.Therefore, in our designs, we make use of Cns3, producing PTN together with COR in order to guarantee a high yield of COR.Previous studies have also proved that the NK domain of Cns3, one of it's two function domains, is responsible for PTN biosynthesis, which is why we only use part of Cns3.
As is mentioned above, this part is an expression cassette of HisG-MF that is half of Cns3 containing the HisG domain. 2019 SCU-China validated its both in expression and function in Saccharomyces cerevisiae.To verify it’s function, we did RT-qPCR to detect the existence of Cns3-MF mRNA. (Figure. 1) As for function validation, we performed HPLC to detect PTN, the catalysate of Cns3-MF(Figure. 2). And we successfully demonstrated the cns3-mf transcription and function finally.
Figure. 1 RT-qPCR of Cns3-HisG-mf\Cns3-HisG-only.
No-load is the S. cerevisiae without transformed plasmids. The reference gene is PGK1. The RNA of cns3 is detected successfully. The bars are SE.
Figure. 2 HPLC analysis of PTN
a. Top: Standard sample of 5mM PTN.
Bottom: Supernatant fraction of Cns3-hisG-MF sample. The strain BY4741 was culturing in YPDG media for 3 days.
b. Top: Standard sample of 5mM PTN.
Bottom: Supernatant fraction of Cns3-hisG ONLY sample. The strain BY4741 was culturing in YPDG media for 3 days.
c. Top: Standard sample of 5mM PTN.
Bottom: Supernatant fraction of BY4741 sample. The strain BY4741 was culturing in YPDG media for 3 days.