Difference between revisions of "Part:BBa K2997012"
(→Characterization) |
|||
| (5 intermediate revisions by 2 users not shown) | |||
| Line 5: | Line 5: | ||
===Characterization=== | ===Characterization=== | ||
| − | This year, we characterized FNR promoter ([[Part:BBa_K1123000|BBa_K1123000]]) which is an anaerobic promoter. Originally, we used this promoter to drive the expression of TAL constructs. However, finding that this promoter has a higher expression level of GFP under aerobic conditions, we did not use this promoter for our project. Instead, we used the strong constitutive promoter J23100 as our promoter. | + | This year, we characterized FNR promoter ([[Part:BBa_K1123000|BBa_K1123000]]) which is an anaerobic promoter. Originally, we used this promoter to drive the expression of TAL constructs. However, finding that this promoter has a higher expression level of GFP under aerobic conditions, we did not use this promoter for our project. Instead, we used the strong constitutive promoter [[Part:BBa_J23100 | J23100]] as our promoter. |
| − | We first cultured DH5a pSB1C3-P<sub> | + | We first cultured DH5a pSB1C3-<i>P<sub>fnr</sub></i>-GFP under aerobic and anaerobic conditions. After 10 hours of incubation, we fixed the <i>E. coli</i> cells in 2% agarose gel and placed it on a glass slide, then place the glass slide under a microscope for image capturing. |
<html> | <html> | ||
| Line 16: | Line 16: | ||
<br> | <br> | ||
</html> | </html> | ||
| − | Fig. 1 Differential interference contrast (DIC) microscope image and fluorescence microscope image for a single <i>E. coli</i> cell. | + | Fig. 1. Differential interference contrast (DIC) microscope image and fluorescence microscope image for a single <i>E. coli</i> cell. |
We then compared the brightness of a single <i>E. coli</i> cell under a fluorescence microscope using ImageJ Intensity Processing. The results are shown below. | We then compared the brightness of a single <i>E. coli</i> cell under a fluorescence microscope using ImageJ Intensity Processing. The results are shown below. | ||
| Line 28: | Line 28: | ||
</html> | </html> | ||
Fig. 2. Quantification of single <i>E. coli</i> cell (n=3) fluorescence intensity using ImageJ Intensity Processing after 10 hours of incubation in both aerobic and anaerobic conditions. | Fig. 2. Quantification of single <i>E. coli</i> cell (n=3) fluorescence intensity using ImageJ Intensity Processing after 10 hours of incubation in both aerobic and anaerobic conditions. | ||
| + | |||
| + | As seen in Fig. 2, after 10 hours of incubation, the fluorescence signal in single <i>E. coli</i> cell is significantly higher in aerobic conditions than in anaerobic conditions. Literature has reported that GFP requires oxygen molecules to fold properly<sup>[1]</sup> before it can emit fluorescence signal. We cannot exclude the possibility that in anaerobic culture, GFP protein is not folding properly and thus affecting the measurement result. However, we can still conclude this promoter is not anaerobic specific. It is recommended that future iGEMers should use other reporter genes that will not be affected by oxygen like luciferase to further characterize this Biobrick. | ||
| + | |||
| + | ===Refence=== | ||
| + | [1] Coralli, C., Maja Cemazar, Chryso Kanthou, Tozer, G. M., & Dachs, G. U. (2001). Limitations of the Reporter Green Fluorescent Protein under Simulated Tumor Conditions. <i>Cancer Research</i>, 61(12), 4784–4790. | ||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 15:31, 21 October 2019
Pfnr-GFP
Characterization
This year, we characterized FNR promoter (BBa_K1123000) which is an anaerobic promoter. Originally, we used this promoter to drive the expression of TAL constructs. However, finding that this promoter has a higher expression level of GFP under aerobic conditions, we did not use this promoter for our project. Instead, we used the strong constitutive promoter J23100 as our promoter.
We first cultured DH5a pSB1C3-Pfnr-GFP under aerobic and anaerobic conditions. After 10 hours of incubation, we fixed the E. coli cells in 2% agarose gel and placed it on a glass slide, then place the glass slide under a microscope for image capturing.
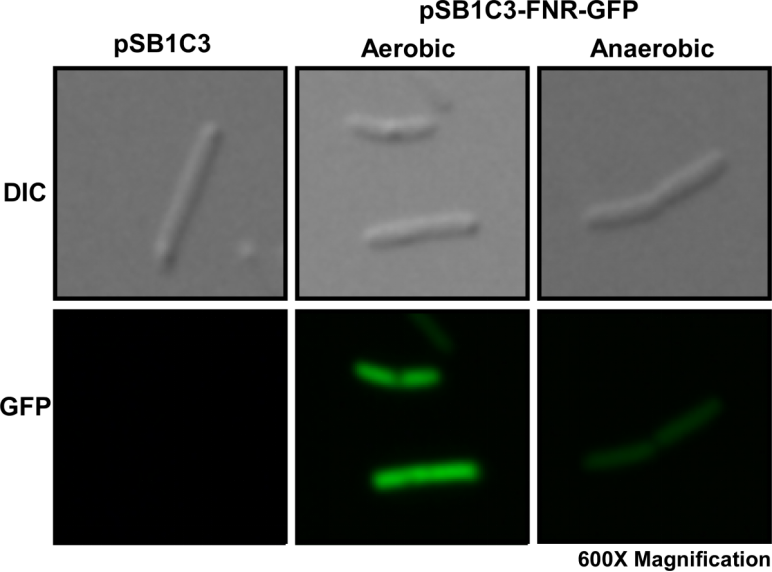
Fig. 1. Differential interference contrast (DIC) microscope image and fluorescence microscope image for a single E. coli cell.
We then compared the brightness of a single E. coli cell under a fluorescence microscope using ImageJ Intensity Processing. The results are shown below.

Fig. 2. Quantification of single E. coli cell (n=3) fluorescence intensity using ImageJ Intensity Processing after 10 hours of incubation in both aerobic and anaerobic conditions.
As seen in Fig. 2, after 10 hours of incubation, the fluorescence signal in single E. coli cell is significantly higher in aerobic conditions than in anaerobic conditions. Literature has reported that GFP requires oxygen molecules to fold properly[1] before it can emit fluorescence signal. We cannot exclude the possibility that in anaerobic culture, GFP protein is not folding properly and thus affecting the measurement result. However, we can still conclude this promoter is not anaerobic specific. It is recommended that future iGEMers should use other reporter genes that will not be affected by oxygen like luciferase to further characterize this Biobrick.
Refence
[1] Coralli, C., Maja Cemazar, Chryso Kanthou, Tozer, G. M., & Dachs, G. U. (2001). Limitations of the Reporter Green Fluorescent Protein under Simulated Tumor Conditions. Cancer Research, 61(12), 4784–4790.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 39
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 790
