Difference between revisions of "Part:BBa K3168007"
CMichielsen (Talk | contribs) |
EvaHanckmann (Talk | contribs) |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 2: | Line 2: | ||
===dCas9-BRET=== | ===dCas9-BRET=== | ||
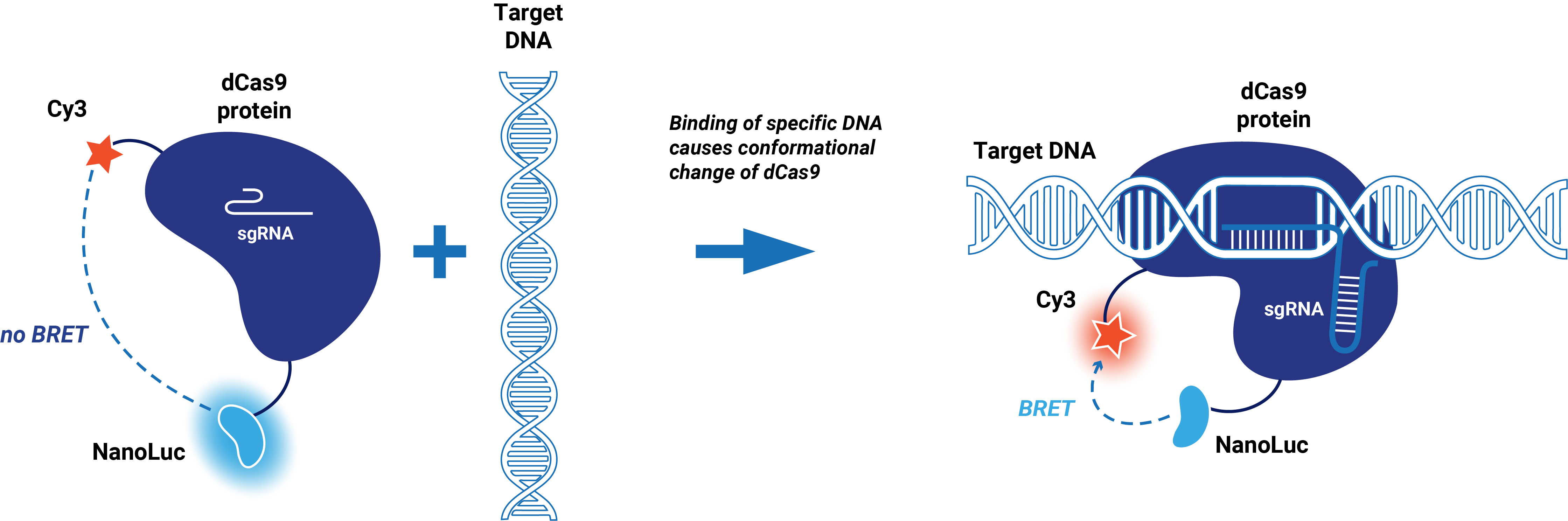
| − | This composite part is made up of two basic parts. The first basic part (BBa_K3168001) codes for a catalytically dead CRISPR associated protein 9 (dCas9) which is circularly | + | This composite part is made up of two basic parts. The first basic part [https://parts.igem.org/wiki/index.php?title=Part:BBa_K3168001 (BBa_K3168001)] codes for a catalytically dead CRISPR associated protein 9 (dCas9) which is circularly permuted and only contains one cysteine (S867C) at the HNH domain to enable dye incorporation via maleimide coupling (Sternberg, 2015). dCas9 binds to a specific target on double-stranded DNA. This specific target is determined by the guide RNA, which makes the target sequence of this protein very modular. The second basic part [https://parts.igem.org/wiki/index.php?title=Part:BBa_K3168006 (BBa_K3168006)], which is fused to dCas9, consists of an (SGG)<sub>2</sub> linker and cysteine-free NanoLuc. A Cy3 dye can be incorporated at S867C via maleimide coupling. |
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | When a Cy3 dye is incorporated, the conformational change of the HNH domain when dCas9 binds to DNA can be detected. In the presence of the substrate (Furimazine), blue light is emitted by NanoLuc. The emission spectrum of NanoLuc overlaps with the excitation spectrum of Cy3. When this fusion protein binds to its target, a conformational change occurs which brings the Cy3 dye close to NanoLuc. This enables bioluminescent resonance energy transfer (BRET), which causes the emission of red light. | + | When a Cy3 dye is incorporated, the conformational change of the HNH domain when dCas9 binds to DNA can be detected. In the presence of the substrate (Furimazine), blue light is emitted by NanoLuc. The emission spectrum of NanoLuc overlaps with the excitation spectrum of Cy3. When this fusion protein binds to its target, a conformational change occurs which brings the Cy3 dye close to NanoLuc. This enables bioluminescent resonance energy transfer (BRET), which causes the emission of red light (Figure 1). |
[[File:T--TU_Eindhoven--dCas9-BRET.png|700px|]] | [[File:T--TU_Eindhoven--dCas9-BRET.png|700px|]] | ||
Latest revision as of 15:26, 21 October 2019
dCas9-BRET
This composite part is made up of two basic parts. The first basic part (BBa_K3168001) codes for a catalytically dead CRISPR associated protein 9 (dCas9) which is circularly permuted and only contains one cysteine (S867C) at the HNH domain to enable dye incorporation via maleimide coupling (Sternberg, 2015). dCas9 binds to a specific target on double-stranded DNA. This specific target is determined by the guide RNA, which makes the target sequence of this protein very modular. The second basic part (BBa_K3168006), which is fused to dCas9, consists of an (SGG)2 linker and cysteine-free NanoLuc. A Cy3 dye can be incorporated at S867C via maleimide coupling.
Usage and Biology
When a Cy3 dye is incorporated, the conformational change of the HNH domain when dCas9 binds to DNA can be detected. In the presence of the substrate (Furimazine), blue light is emitted by NanoLuc. The emission spectrum of NanoLuc overlaps with the excitation spectrum of Cy3. When this fusion protein binds to its target, a conformational change occurs which brings the Cy3 dye close to NanoLuc. This enables bioluminescent resonance energy transfer (BRET), which causes the emission of red light (Figure 1).
Figure 1. Schematic representation dCas9-BRET system.
References
Oakes, B. L., Fellmann, C., Rishi, H., Taylor, K. L., Ren, S. M., Nadler, D. C., ... & Savage, D. F. (2019). CRISPR-Cas9 circular permutants as programmable scaffolds for genome modification. Cell, 176(1-2), 254-267.
Sternberg, S. H., LaFrance, B., Kaplan, M., & Doudna, J. A. (2015). Conformational control of DNA target cleavage by CRISPR–Cas9. Nature, 527(7576), 110.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 2143
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 261
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]