Difference between revisions of "Part:BBa K1104201"
| Line 53: | Line 53: | ||
<li> <b>Sample B</b> - <i>E.coli</i> BL21(DE3) cells without engineered plasmid. </li> | <li> <b>Sample B</b> - <i>E.coli</i> BL21(DE3) cells without engineered plasmid. </li> | ||
</ul> | </ul> | ||
| − | |||
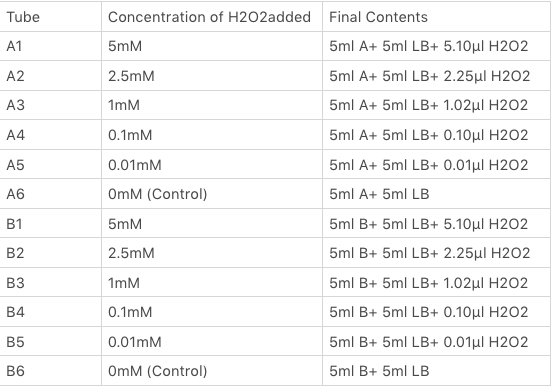
We divided our 50ml test tubes of Sample A and B culture into twelve 15ml test tubes, each contained 5ml culture broth and 5ml LB with OD600 0.4. H2O2 was added to 10 of the tubes in the following concentrations: | We divided our 50ml test tubes of Sample A and B culture into twelve 15ml test tubes, each contained 5ml culture broth and 5ml LB with OD600 0.4. H2O2 was added to 10 of the tubes in the following concentrations: | ||
<img src="https://2019.igem.org/wiki/images/1/15/T--SUIS_Shanghai--H2O2conc.png" height=350px width=500px align=center> | <img src="https://2019.igem.org/wiki/images/1/15/T--SUIS_Shanghai--H2O2conc.png" height=350px width=500px align=center> | ||
| − | |||
| − | |||
| − | |||
<br> | <br> | ||
<p>After 2 hours of incubation, we extracted 1.4 ml culture from each of tubes and placed in twelve 1.5ml sterile centrifuge tube and centrifuged at 4000rpm for 4 minutes. After removing the supernatant, we washed the cell with PBS buffer. Then, 200 μM culture of A1, A2, A3, A4, A5, A6, B1, B2, B3, B4, B5, B6 was added into 96 well white polystyrene microplate and black polystyrene microplate, each with three samples. We measured the OD600 and Fluorescence (Excitation: 485nm/ Emission: 528nm) by using plate reader. The data was recorded. After that, we calculated the average OD600 and Fluorescence for all samples. For each of samples, we divided the relative fluorescence value (RFV) by the average OD600.</p> | <p>After 2 hours of incubation, we extracted 1.4 ml culture from each of tubes and placed in twelve 1.5ml sterile centrifuge tube and centrifuged at 4000rpm for 4 minutes. After removing the supernatant, we washed the cell with PBS buffer. Then, 200 μM culture of A1, A2, A3, A4, A5, A6, B1, B2, B3, B4, B5, B6 was added into 96 well white polystyrene microplate and black polystyrene microplate, each with three samples. We measured the OD600 and Fluorescence (Excitation: 485nm/ Emission: 528nm) by using plate reader. The data was recorded. After that, we calculated the average OD600 and Fluorescence for all samples. For each of samples, we divided the relative fluorescence value (RFV) by the average OD600.</p> | ||
Revision as of 15:07, 21 October 2019
TrxC promoter
Introduction
TrxCp is a ROS-induced promoter controlled by OxyR (transcription factor) which is activated by ROS (Reactive Oxygen Species).
An intramolecular disulfide bond between Cys199 and Cys208 will form in an OxyR transcription factor, and the active OxyR molecule will bind to the binding site of TrxCp, making the following circuit turned on. In our project: Bee. coli, we use TrxCp as the sensor of Bee. coli to sense Nosema ceranae. After Nosema ceranae enter bees' midgut, the natural immune reaction will lead to ROS release by epithelia of bees' midgut. We select several promoters that can be induced by OxyR (Part:BBa_K1104200), which are activated by ROS. One of these promoters is TrxCp.
OxyR is activator of TrxC promoter. More details about OxyR can be found on the PartRegistry page: Part:BBa_K1104200.
Information of TrxCp on [http://www.ecocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-3104 Ecocyc]
We get information of TrxCp from [http://www.ecocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-3104 Ecocyc],
TrxCp is regulated by σ70, and there are two TFBSs upstream for OxyR. We also did some literature study, according to the citation.
In E. coli genome, thioredoxin (trxC) will express in response to elevated H2O2 level. Expression of genes encoding thioredoxin (trxC) is regulated by OxyR. Oxidized OxyR binds directly to TrxCp and induces its expression.
- Reference: Ritz, D. (January 28, 2000). Thioredoxin 2 Is Involved in the Oxidative Stress Response in Escherichia coli. Journal of Biological Chemistry, 275, 4, 2505-2512.
We strimmed trxC coding out of the sequence on [http://www.ecocyc.org/ECOLI/NEW-IMAGE?type=OPERON&object=TU0-3104 Ecocyc], and only take the part including 2 OxyR TFBSs and TrxCp region.
Usage and Biology
We designed circuit fighting against Nosema ceranae. After Nosema ceranae infected midgut cells of bees, Bee. coli should sense the pathogen first before the following circuit (fighting against Nosema ceranae)is triggered, and substance such as Defensin (Part:BBa_K1104300), Abaesin (Part:BBa_K1104301) (more details on [http://2013.igem.org/Team:NYMU-Taipei/Project/Inhibition/Killing Killing Nosema] page) in the following circuit will express.
Besides, in order to enhance the strength of TrxCp, we added a device (more details on [http://2013.igem.org/Team:NYMU-Taipei/Project/Inhibition/Sensor Sensing Nosema] page).
Contribution - Characterization
SUIS_Shanghai iGEM 2019 provides characterization for this part. We first got the above gene circuit sequence synthesized by Genscript onto expression vector pET30a(+) and transformed into E.coli BL21(DE3) cells. Overnight cultures were later tested for GFP fluorescence. The above construct should only produce GFP in the presence of enough OxyR transcription factor and a large enough concentration of H2O2. OxyR is activated by ROS (Reactive Oxygen Species) which contains a reactive cysteine residue at position 199 (Cys-199). Upon exposure to a ROS, Cys-199 is oxidized resulting in the formation of an intermolecular disulfide bond between Cys-199 and Cys-208. This causes a conformational change, activating the OxyR transcription factor which can bind to regularly sequences and thus positively regulate some genes. As mentioned on this page the TrxC promoter is a regulatory sequence for the thioredoxin (TrxC) gene so after synthesis of the gene circuit we designed an experiment to add experimental characterization of this part by activating the promoter to express GFP. H2O2 was added to cell cultures to induce the expression of GFP by first oxidizing the reactive Cys-199 on OxyR and thus activating this transcription factor for TrxC promoter. This gene construct will express OxyR all the time but downstream genes from TrxC promoter will only be expressed upon binding of active OxyR to the binding regions. OxyR is activated by the oxidation of a reactive cysteine residue (Cys-199) by a reactive oxygen species (ROS).
Experiment
- Sample A - E.coli BL21(DE3) cells containing expression plasmid and the gene sequence above.
- Sample B - E.coli BL21(DE3) cells without engineered plasmid.
After 2 hours of incubation, we extracted 1.4 ml culture from each of tubes and placed in twelve 1.5ml sterile centrifuge tube and centrifuged at 4000rpm for 4 minutes. After removing the supernatant, we washed the cell with PBS buffer. Then, 200 μM culture of A1, A2, A3, A4, A5, A6, B1, B2, B3, B4, B5, B6 was added into 96 well white polystyrene microplate and black polystyrene microplate, each with three samples. We measured the OD600 and Fluorescence (Excitation: 485nm/ Emission: 528nm) by using plate reader. The data was recorded. After that, we calculated the average OD600 and Fluorescence for all samples. For each of samples, we divided the relative fluorescence value (RFV) by the average OD600.

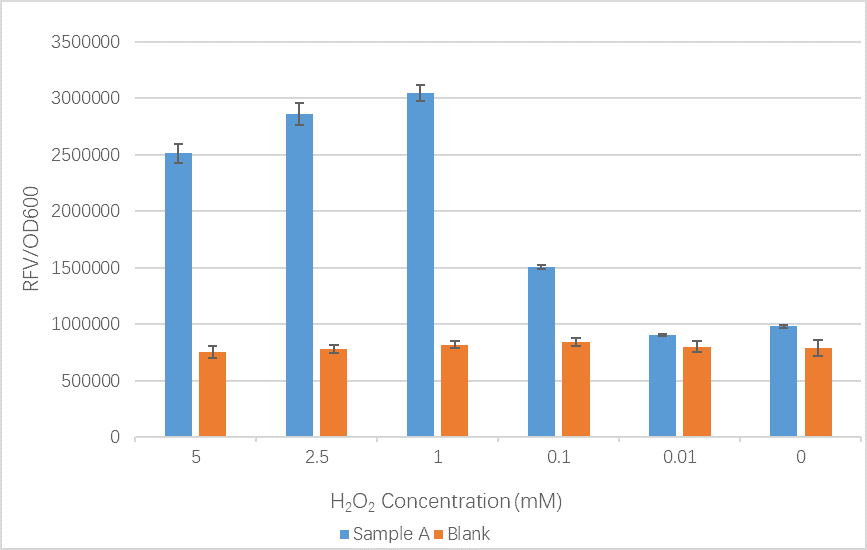
Comparison of relative fluorescence value (RFV)/OD600 of Sample A, with OxyR transcription factor (BBa_K1104200) from iGEM13_NYMU-Taipeiunder and Reactive Oxygen Species induced promoter TrxCp (BBa_K1104201), and Blank (unmodified BL21 E. coli), under six H2O2 concentrations. Results indicate a significant difference between Sample A and Blank in all concentrations measured.
This is the relative fluorescence value (RFV)/OD600, an indication of promoter performance, of sample A and a control group under different H2O2 concentration. Similar to the trend of the original result from team NYMU-Taipei, the RFV/OD600 value tend to increase as the concentration of H2O2 increase, despite the gradual decreasing fluorescence value after the concentration exceed 1mM. Accelerating protein degradation of GFP might be a possible explanation for this distinction. Protein biodegradability is said to increase along with peroxide concentration.
Related Parts
- Part:BBa_K1104241: TrxCp+E0840, reporter with GFP for promoter testing.
- Part:BBa_K1104202: HemHp, another OxyR-induced promoter.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]






