Difference between revisions of "Part:BBa K3030017"
| Line 31: | Line 31: | ||
none | none | ||
| − | [[File:T--XJTLU-CHINA--Figure4.png|thumb|center|600px|'''Figure 3:''' Fig. 2: | + | [[File:T--XJTLU-CHINA--Figure4.png|thumb|center|600px|'''Figure 3:''' Fig. 2: S]] |
| − | [[File:T--XJTLU-CHINA--Figure4.png|thumb|center|700px|'''Figure 3:''' Fig. 2: | + | [[File:T--XJTLU-CHINA--Figure4.png|thumb|center|700px|'''Figure 3:''' Fig. 2: S]] |
| + | |||
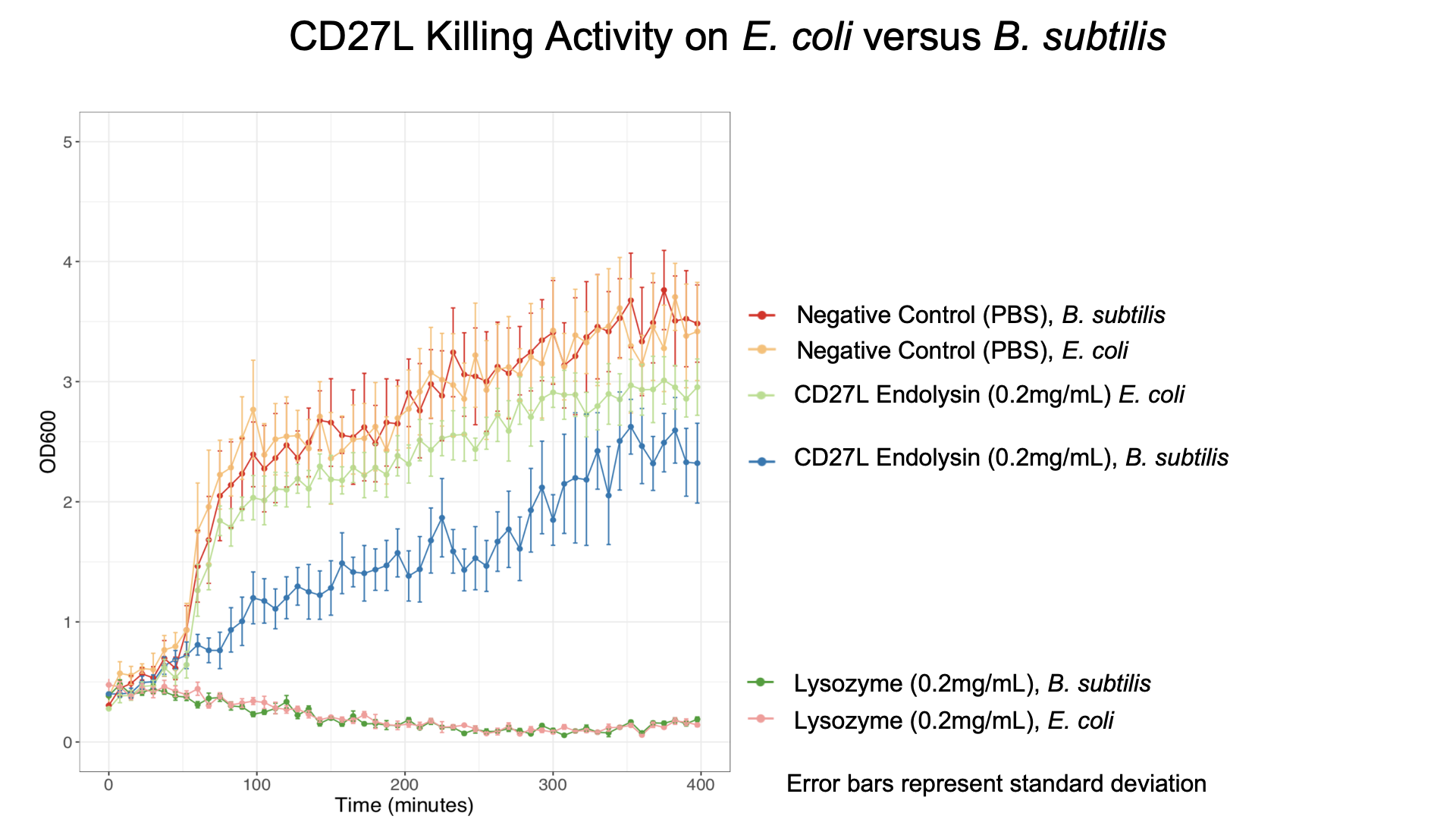
| + | [[File:BBa_K3183200_Bsubtilis.png|thumb|left|430px|'''Figure 6:''' C]] | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Revision as of 12:56, 21 October 2019
theophylline riboswitch with optimized RBS + eGFP
This is a part that can test the function of the optimized RBS. The theophylline riboswitch is initially designed by Wieland, M. and Hartig J. S. (2008). We optimize this sequence by replacing the Shine-Dalgarno (SD) sequence with our calculated RBS by thermodynamic model. This switch is theophylline responsive which means this switch will undergo self-cleavage at the presence of theophylline. After self-cleavaging, ribosomes can bind with mRNA and translation will initiate. We use eGFP(BBa_E0040) as our reporter. The fluorescence intensity/OD. 600 can indicate the functionality of this switch. We place this part at the downstream of pBAD promoter. With the induce of L-arabinose, BL21(DE3) cells will transcribe mRNAs containing theophylline switch and eGFP coding sequence. At the presence of theophylline, eGFP protein will be translated and by detecting the fluorescence intensity/OD. 600, the functionality of this switch can be characterized.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 754
Background
H2
H3
H4
H5
H6
<h7>H7</h7>
none
none