Difference between revisions of "Part:BBa I13522"
Smelissali (Talk | contribs) |
(→Overview:) |
||
| (10 intermediate revisions by 6 users not shown) | |||
| Line 4: | Line 4: | ||
Untagged GFP behind a constitutive promoter. | Untagged GFP behind a constitutive promoter. | ||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | GFP only control | ||
| − | < | + | The version shipped with the 2008 iGEM distribution does not appear to function. Unclear why. [[User:Jkm|Jkm]] 18:40, 22 August 2008 (UTC) |
| + | <br><br> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
<partinfo>BBa_I13522 SequenceAndFeatures</partinfo> | <partinfo>BBa_I13522 SequenceAndFeatures</partinfo> | ||
| Line 18: | Line 19: | ||
==Pictures== | ==Pictures== | ||
| − | + | <gallery> | |
| − | + | Image:I13522 lightbox.jpg|BBa_I13522 visualized under non-UV lightbox | |
| + | Image:I13522 - UV 254nm.JPG|BBa_I13522 visualized under 254nm wavelength UV lightbox | ||
| + | </gallery> | ||
| + | |||
| + | ==Characterization of [[Chassis/Cell-Free_Systems|Cell-Free Chassis]]== | ||
| + | This part was used to characterize the cell-free chassis. Details of characterization can be found on the [[Chassis/Cell-Free Systems/Commercial E.coli S30|cell-free characterization]] page. Other interesting isothermal properties of this construct are available at the [http://2007.igem.org/Imperial/Cell_by_Date/Testing Cell By Date project page] of Imperial 2007 wiki. | ||
| + | {|border=1 width="90%" align="center" | ||
| + | |- | ||
| + | !width="20%" style="background:#CCCCFF"|Parameter | ||
| + | !width="80%"|Value and Description | ||
| + | |-align="center" | ||
| + | |style="background:#EEEEFF"|Optimal Temperature | ||
| + | |Expression increases with temperature from 4°C to 30°C | ||
| + | |-align="center" | ||
| + | |style="background:#EEEEFF"|Optimal DNA Concentration | ||
| + | |Works optimally with 2µg of DNA in 60 µl of cell-extract | ||
| + | |-align="center" | ||
| + | |style="background:#EEEEFF"|Peak Expression Time | ||
| + | |Peak rate of output occurs after approximately 85 minutes | ||
| + | |-align="center" | ||
| + | |style="background:#EEEEFF"|Expression Lifespan | ||
| + | |Expression levels off after 2 hours | ||
| + | |-align="center" | ||
| + | |style="background:#EEEEFF"|Expression Capacity | ||
| + | |Can synthesize over 200pmols of GFPmut3b in 6 hours | ||
| + | |} | ||
| + | |||
| + | ='''IISER Kolkata 2019'''= | ||
| + | |||
| + | === The Characterization of BBa_K381001 === | ||
| + | |||
| + | <p> | ||
| + | In our project to combat Leishmaniasis using a genetically modified | ||
| + | bacterium we planned to use bacterial promoter pTET as a upstream of our | ||
| + | coding region but for it to act effectively we need to quantify that its | ||
| + | promoter activity is comparable in both bacterial and mammalian culture | ||
| + | medium. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Aim</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | BBa_I13522 was used as a test circuit to compare the activity of pTET | ||
| + | bacterial promoter in bacterial medium Luria Broth and mammalian cell | ||
| + | culture medium DMEM. | ||
| + | </p> | ||
| + | <p> | ||
| + | <strong>Method</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | E. coli DH5α were transformed using re-suspended BBa_I13522 pSB1C3 plasmids | ||
| + | from iGEM 2019 distribution kit and selected using Chloramphenicol LB agar | ||
| + | plates. | ||
| + | </p> | ||
| + | <ol> | ||
| + | <li> | ||
| + | Single positive colony of BBa_I13522 was added to 10ml of LB media | ||
| + | and left overnight at 37° C on 150 rpm in an incubator. Along with | ||
| + | this, a positive control with non-transformed E coli cells was also | ||
| + | kept in liquid culture in the similar conditions. | ||
| + | </li> | ||
| + | <li> | ||
| + | 100 µL of overnight culture was added to 10ml of LB media and DMEM | ||
| + | media in two different test tubes. For positive control secondary | ||
| + | culture was added in 10ml of LB media and all the test tubes were | ||
| + | left in incubator for 4 hours until the OD for the control reached | ||
| + | 0.6 | ||
| + | </li> | ||
| + | <li> | ||
| + | 2 mL of liquid culture from each of the test tube was centrifuged, | ||
| + | washed and re-suspended in 500 µL of Phosphate-Buffered Saline | ||
| + | (PBS) | ||
| + | </li> | ||
| + | <li> | ||
| + | 100uL of the re-suspended culture was added to each well of 96 well | ||
| + | plate. | ||
| + | </li> | ||
| + | <li> | ||
| + | Varioskan LUX multimode reader was used to measure the GFP | ||
| + | fluorescence: Excitation at 494 nm and emission at 525 nm. | ||
| + | </li> | ||
| + | </ol> | ||
| + | <p> | ||
| + | <strong>Result</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | BBa_I13522 transformed <em>E. coli</em> shows similar GFP expression in | ||
| + | both bacterial and mammalian media. | ||
| + | </p> | ||
| + | https://2019.igem.org/wiki/images/5/53/T--IISER_Kolkata--ptet_gfp.JPG | ||
| + | |||
| + | Mean GFP fluorescence (Excitation 494nm/Emission 525nm) of BBa_I13522 transformed E.coli grown in two different media as compared untransformed E. coli grown in LB media and suspended in PBS | ||
| + | <p> | ||
| + | <strong>Conclusion</strong> | ||
| + | </p> | ||
| + | <p> | ||
| + | The bacterial promoter pTET constitutively expresses comparable amount the | ||
| + | downstream coding region (GFP in this case) irrespective of medium in which | ||
| + | it is grown. | ||
| + | </p> | ||
| + | |||
| + | <br/> | ||
| + | =UIUC_Illinois 2019 Characterization= | ||
| + | ==Overview:== | ||
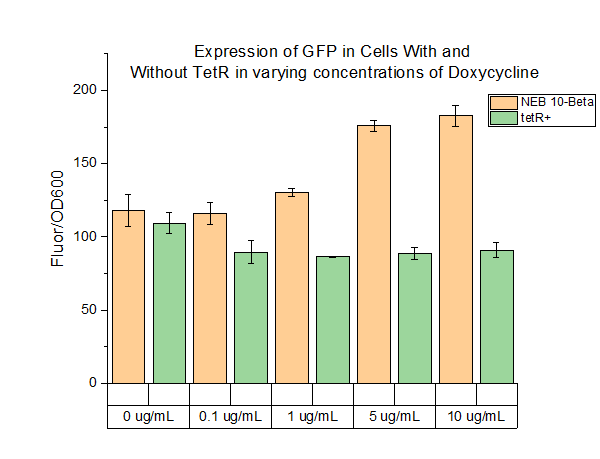
| + | We characterized this part using fluorescence at different concentrations of doxycycline (DOX) in order to determine the level of DOX we would need to use to induce genes in the system we were testing. The plasmid was transformed into two cell types: NEB 10-beta and a cell strain containing tetR. Cell cultures were grown up overnight in 5 mL of LB with chloramphenicol at 37 C. The folloTwing day, 990 uL of LB, 10 uL of cell culture, and 1 uL of chloramphenicol were added to 1.5 mL Eppendorf tubes. The tubes were incubated for 4 hours at 37 C. After 4 hours, DOX of varying concentrations was added to the tubes. Five different conditions were tested, with each condition tested with 4 replicates. The tubes were then incubated at 37 C for 16 hours. 200 uL aliquots were then taken from each tube and spun down for 3 minutes. The cells were resuspended in 200 uL of PBS buffer. 100 uL of resuspended cells were plated on a clear 96 well plate and fluorescence and OD600 were measured. The fluorescence settings were excitation set 485 nm, emission cutoff at 495 nm, and emission at 525 nm. | ||
| + | <br/> | ||
| + | [[File:T--UIUC_Illinois--ptet_fluor_data.png|center]] | ||
| + | <br/> | ||
| + | The above figure shows the measured fluorescence of GFP in NEB 10-Beta normalized by OD600. The addition of DOX at the indicated levels had a slight toxic effect on the cell, resulting a decrease in OD600. This accounts for the increase in fluorescence in NEB 10-Beta cells as concentration increased. The same effect was not seen in the tetR+ cell line, however, as fluorescence levels remained about constant at all concentrations of added DOX. | ||
Latest revision as of 05:58, 21 October 2019
pTet GFP
Untagged GFP behind a constitutive promoter.
Usage and Biology
GFP only control
The version shipped with the 2008 iGEM distribution does not appear to function. Unclear why. Jkm 18:40, 22 August 2008 (UTC)
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 724
Pictures
Characterization of Cell-Free Chassis
This part was used to characterize the cell-free chassis. Details of characterization can be found on the cell-free characterization page. Other interesting isothermal properties of this construct are available at the [http://2007.igem.org/Imperial/Cell_by_Date/Testing Cell By Date project page] of Imperial 2007 wiki.
| Parameter | Value and Description |
|---|---|
| Optimal Temperature | Expression increases with temperature from 4°C to 30°C |
| Optimal DNA Concentration | Works optimally with 2µg of DNA in 60 µl of cell-extract |
| Peak Expression Time | Peak rate of output occurs after approximately 85 minutes |
| Expression Lifespan | Expression levels off after 2 hours |
| Expression Capacity | Can synthesize over 200pmols of GFPmut3b in 6 hours |
IISER Kolkata 2019
The Characterization of BBa_K381001
In our project to combat Leishmaniasis using a genetically modified bacterium we planned to use bacterial promoter pTET as a upstream of our coding region but for it to act effectively we need to quantify that its promoter activity is comparable in both bacterial and mammalian culture medium.
Aim
BBa_I13522 was used as a test circuit to compare the activity of pTET bacterial promoter in bacterial medium Luria Broth and mammalian cell culture medium DMEM.
Method
E. coli DH5α were transformed using re-suspended BBa_I13522 pSB1C3 plasmids from iGEM 2019 distribution kit and selected using Chloramphenicol LB agar plates.
- Single positive colony of BBa_I13522 was added to 10ml of LB media and left overnight at 37° C on 150 rpm in an incubator. Along with this, a positive control with non-transformed E coli cells was also kept in liquid culture in the similar conditions.
- 100 µL of overnight culture was added to 10ml of LB media and DMEM media in two different test tubes. For positive control secondary culture was added in 10ml of LB media and all the test tubes were left in incubator for 4 hours until the OD for the control reached 0.6
- 2 mL of liquid culture from each of the test tube was centrifuged, washed and re-suspended in 500 µL of Phosphate-Buffered Saline (PBS)
- 100uL of the re-suspended culture was added to each well of 96 well plate.
- Varioskan LUX multimode reader was used to measure the GFP fluorescence: Excitation at 494 nm and emission at 525 nm.
Result
BBa_I13522 transformed E. coli shows similar GFP expression in both bacterial and mammalian media.
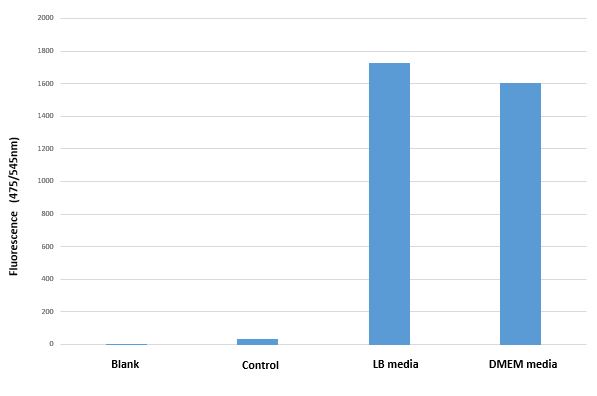
Mean GFP fluorescence (Excitation 494nm/Emission 525nm) of BBa_I13522 transformed E.coli grown in two different media as compared untransformed E. coli grown in LB media and suspended in PBS
Conclusion
The bacterial promoter pTET constitutively expresses comparable amount the downstream coding region (GFP in this case) irrespective of medium in which it is grown.
UIUC_Illinois 2019 Characterization
Overview:
We characterized this part using fluorescence at different concentrations of doxycycline (DOX) in order to determine the level of DOX we would need to use to induce genes in the system we were testing. The plasmid was transformed into two cell types: NEB 10-beta and a cell strain containing tetR. Cell cultures were grown up overnight in 5 mL of LB with chloramphenicol at 37 C. The folloTwing day, 990 uL of LB, 10 uL of cell culture, and 1 uL of chloramphenicol were added to 1.5 mL Eppendorf tubes. The tubes were incubated for 4 hours at 37 C. After 4 hours, DOX of varying concentrations was added to the tubes. Five different conditions were tested, with each condition tested with 4 replicates. The tubes were then incubated at 37 C for 16 hours. 200 uL aliquots were then taken from each tube and spun down for 3 minutes. The cells were resuspended in 200 uL of PBS buffer. 100 uL of resuspended cells were plated on a clear 96 well plate and fluorescence and OD600 were measured. The fluorescence settings were excitation set 485 nm, emission cutoff at 495 nm, and emission at 525 nm.
The above figure shows the measured fluorescence of GFP in NEB 10-Beta normalized by OD600. The addition of DOX at the indicated levels had a slight toxic effect on the cell, resulting a decrease in OD600. This accounts for the increase in fluorescence in NEB 10-Beta cells as concentration increased. The same effect was not seen in the tetR+ cell line, however, as fluorescence levels remained about constant at all concentrations of added DOX.