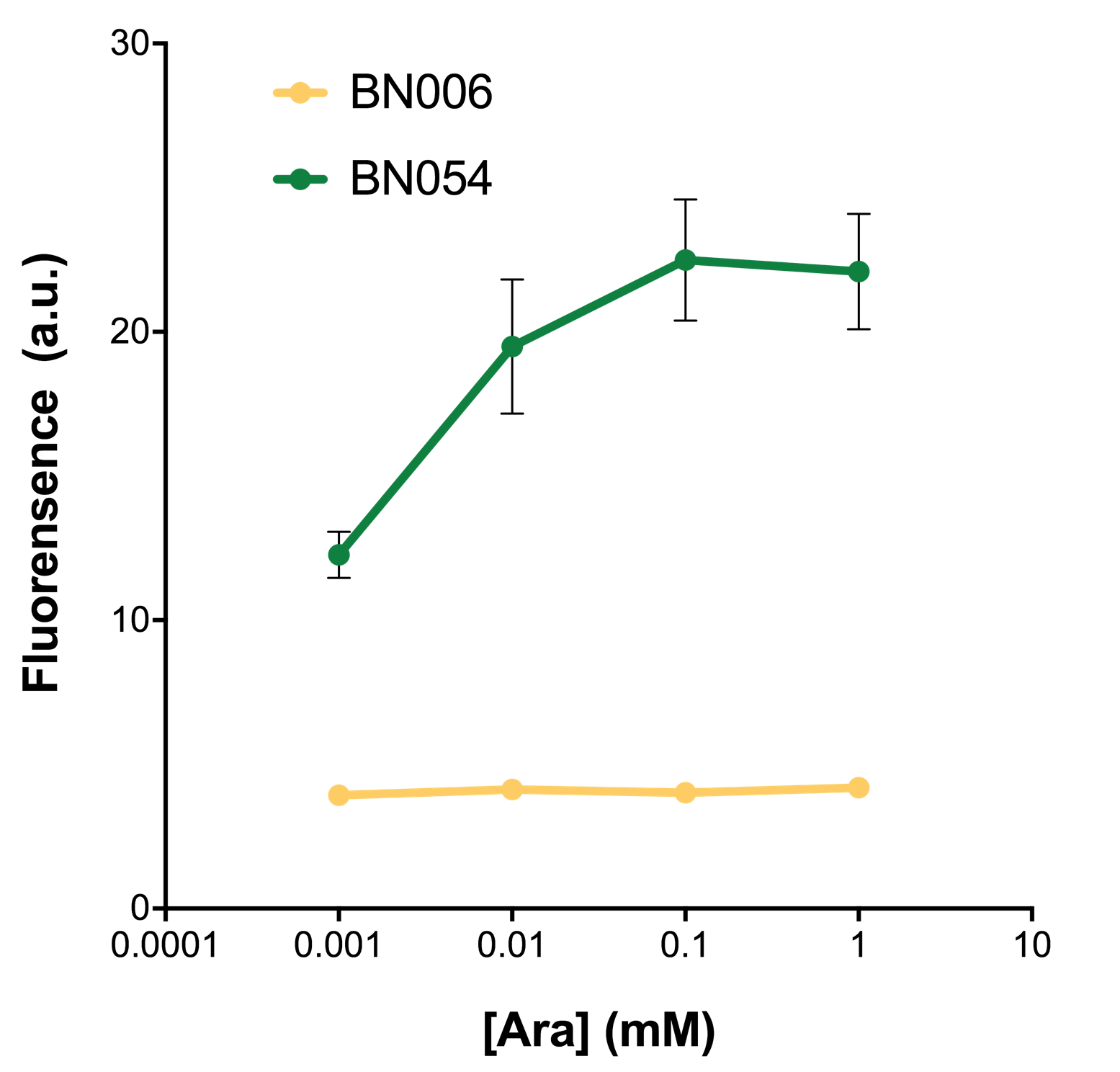Difference between revisions of "Part:BBa K3202029"
Katherine Z4 (Talk | contribs) |
|||
| (63 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K3202029 short</partinfo> | <partinfo>BBa_K3202029 short</partinfo> | ||
| Line 5: | Line 4: | ||
This composite part is designed as a genetic circuit that is used as a control group of our later experiment. | This composite part is designed as a genetic circuit that is used as a control group of our later experiment. | ||
| − | This acts as a control group in our experiment. It is compared to BBa_K3202030 and BBa_K3202031. | + | This acts as a control group in our experiment. It is compared to BBa_K3202030 and BBa_K3202031 in the leakage/inducer module, and later used in the bistable system module. |
| + | |||
| + | =<b>Leakage/Inducer Module</b>= | ||
| + | |||
| + | <hr style="height:0px;border:none;border-top:none solid #555555;" /> | ||
| + | |||
| + | === I.Background === | ||
| + | |||
| + | Gene expression depends largely on transcription and translation. Such processes display activated/non-activated states. For some situations, we would expect zero expression of GOI without the presence of inducer. However, leakage still exist which is due to the incomplete OFF stage of the transcriptional system. Even without proper inducers, GOI is still partly expressed in some case. Such leakage is called the basal leakage.In some cases, as long as one molecule of toxin protein, is translated, it can kill the entire cell. Thus, preventing leakage is a vital in regulating gene expression. Current methods of solving leakage problems includes introducing strong inducible promoters to the system. However, there are substantially no regulating system or transcriptional factor that can function as a technically ON/OFF switch to prevent basal level leakage. This may due to weak RNAP-DNA interactions or different transcriptional capacities and induction rate of different promoters and inducers. | ||
| + | |||
| + | === II.Design === | ||
| + | |||
| + | We mainly focused on two types of inducible transcriptional promoter—Xyls and pBAD. | ||
| + | The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design). | ||
| + | The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The pBAD promoter exhibits an almost all-or-none behavior upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Based on these research findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD. | ||
| + | |||
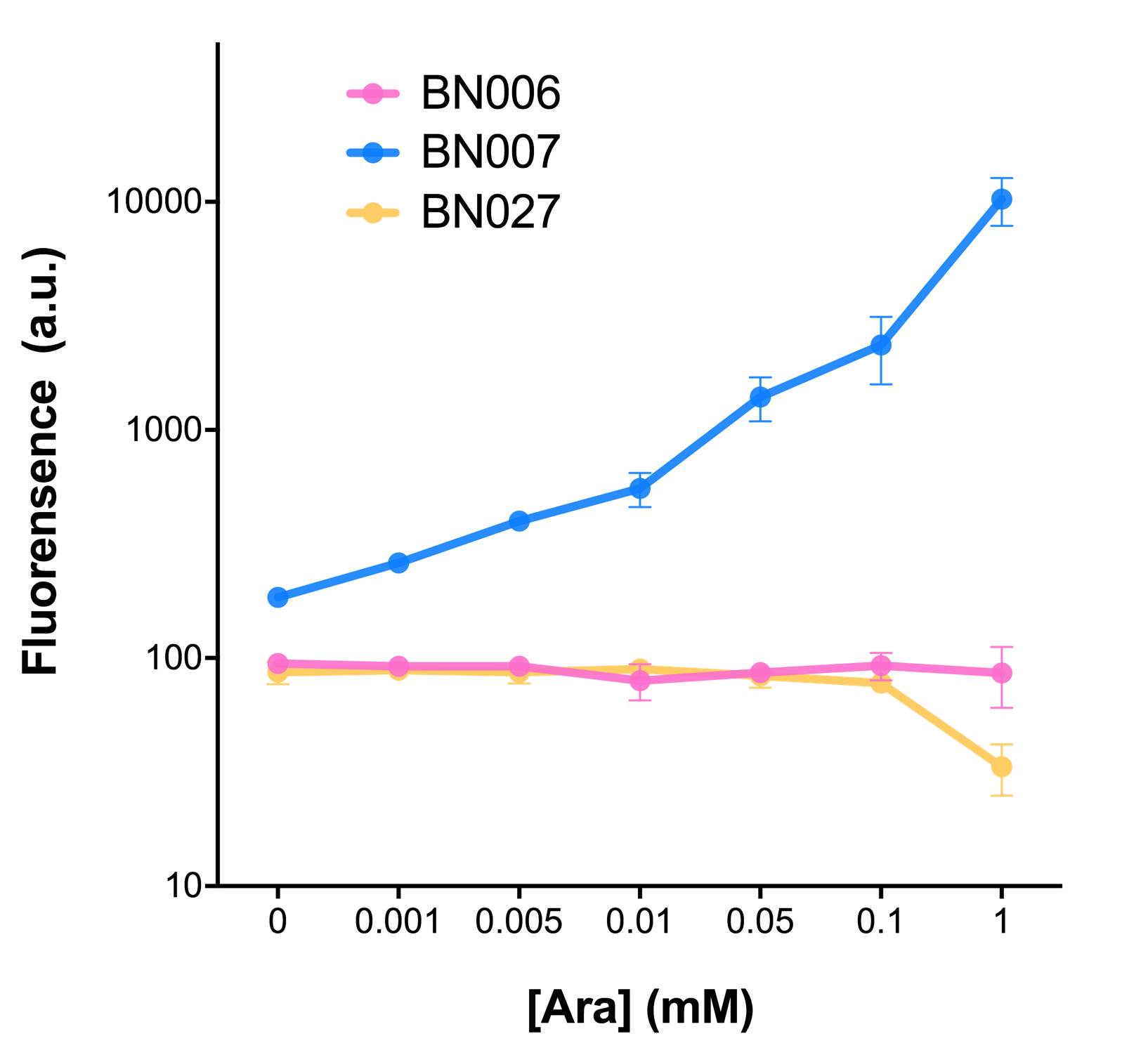
| + | As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006/Contains BBa_K3202029) which suggest the two inducible transcription system function properly. | ||
| + | |||
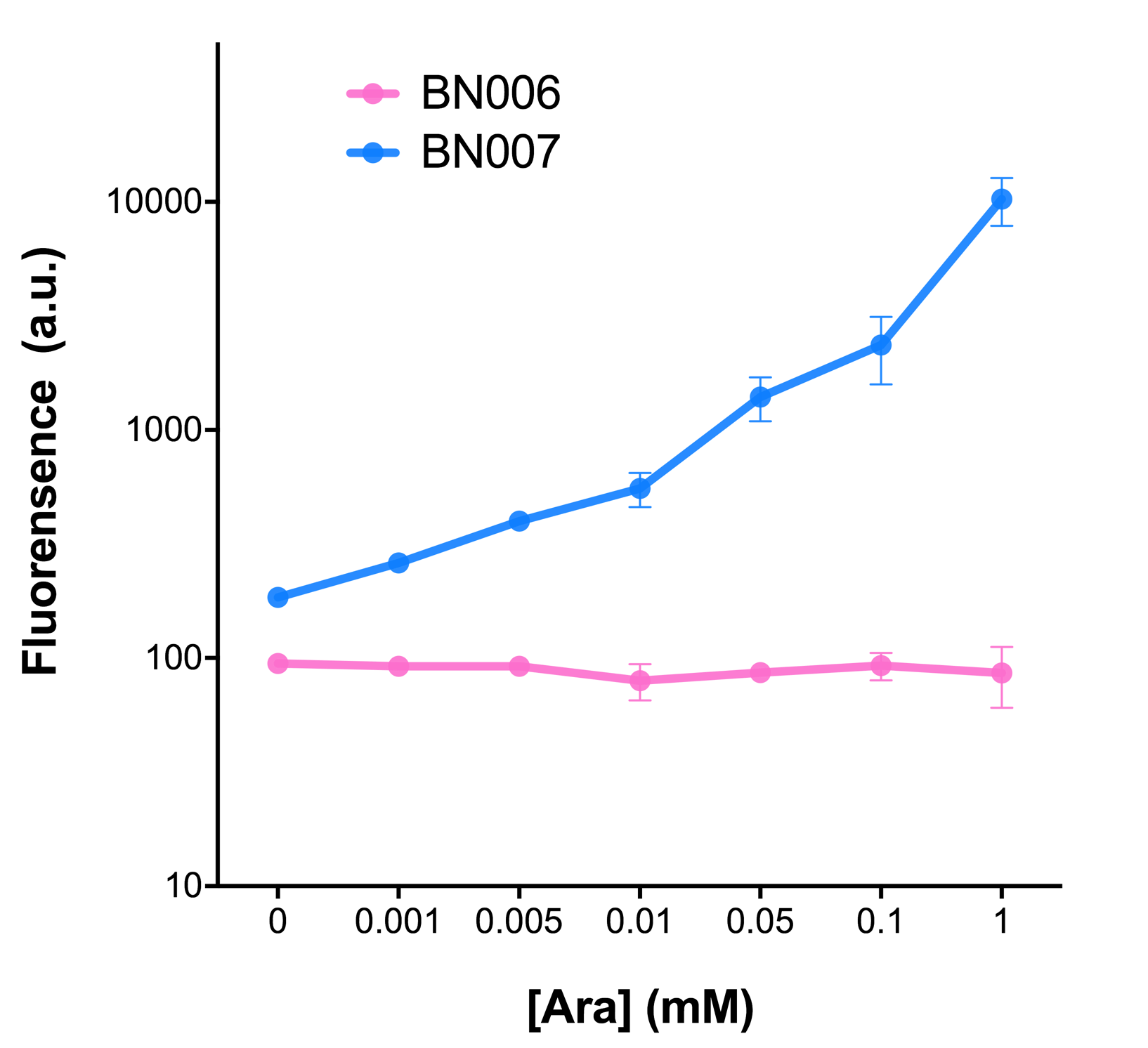
| + | [[File:T--BHSF_ND--Leakage experiment 1.png|429px|thumb|left|alt=Comparison between BN006/Contains BBa_K3202029 and BN007/Contains BBa_K3202030|Figure 1: pBAD(BN007/Contains BBa_K3202030) induction curve at different concentrations after 4 hours compared to backbone.(BN006/Contains BBa_K3202029)]] | ||
| + | |||
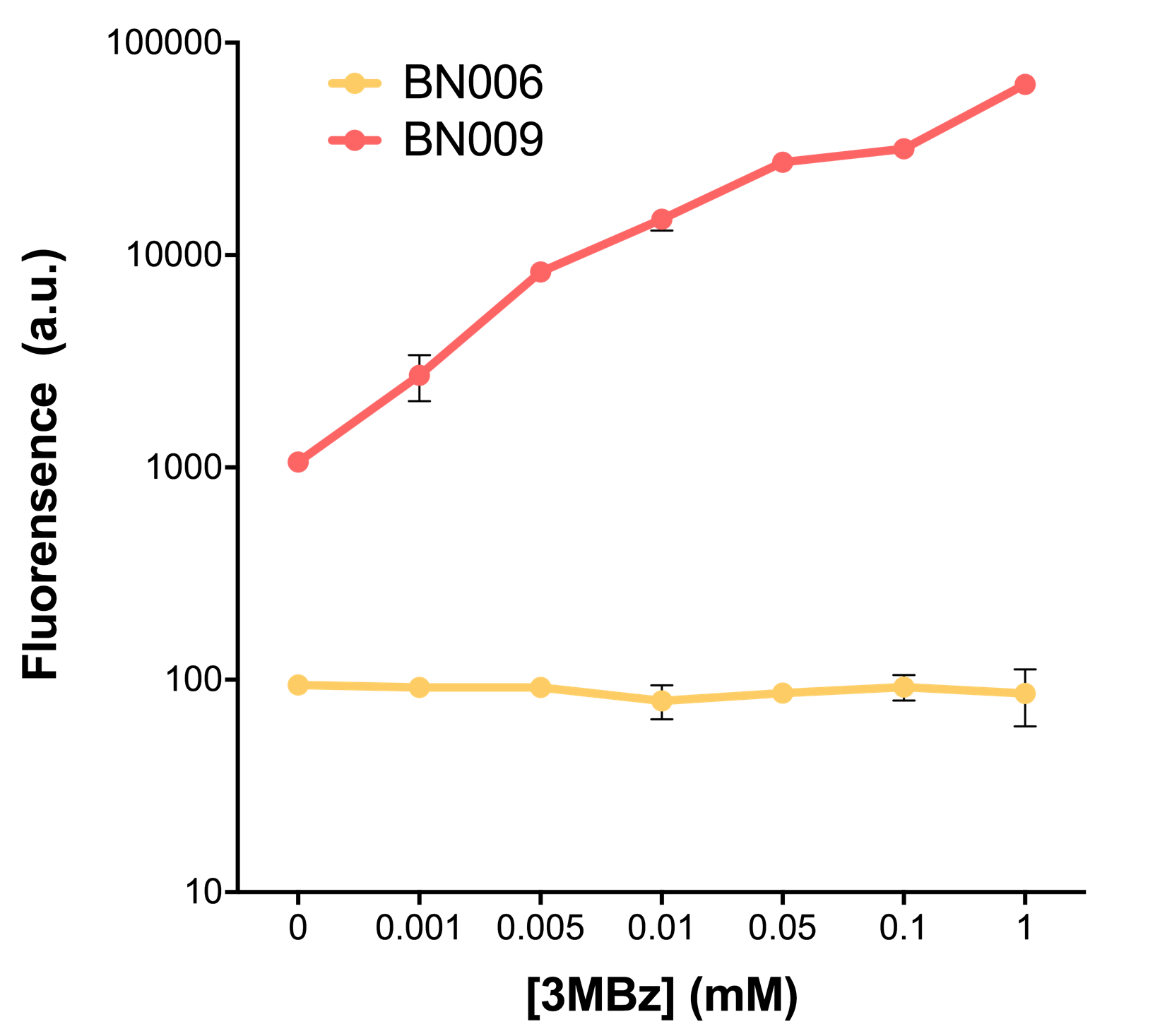
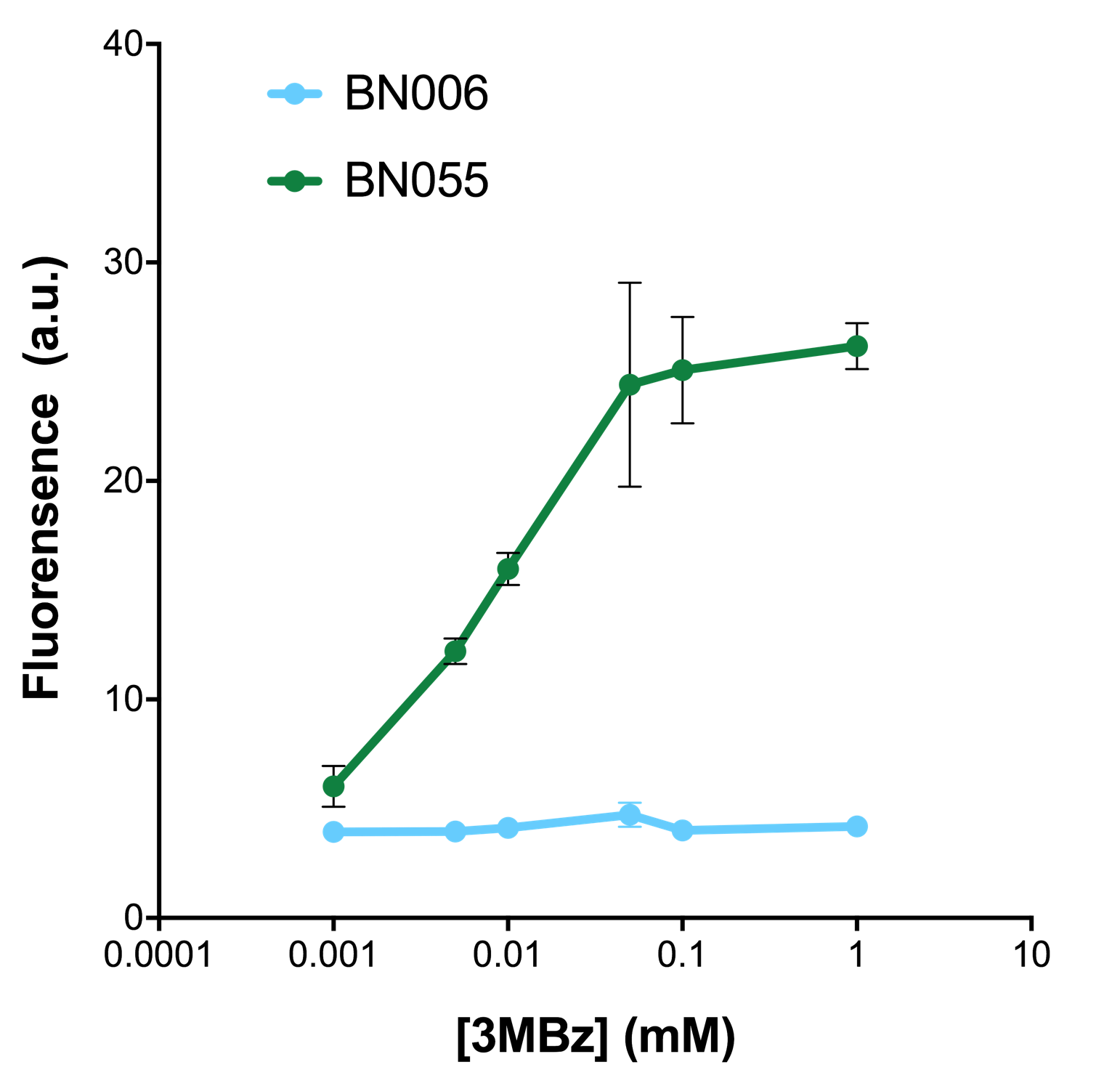
| + | [[File:T--BHSF_ND--Leakage experiment 2.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure 2: Xyls(BN009/Contains BBa_K3202031) induction curve at different concentrations after 4 hours compared to backbone(BN006/Contains BBa_K3202029)]] | ||
| + | |||
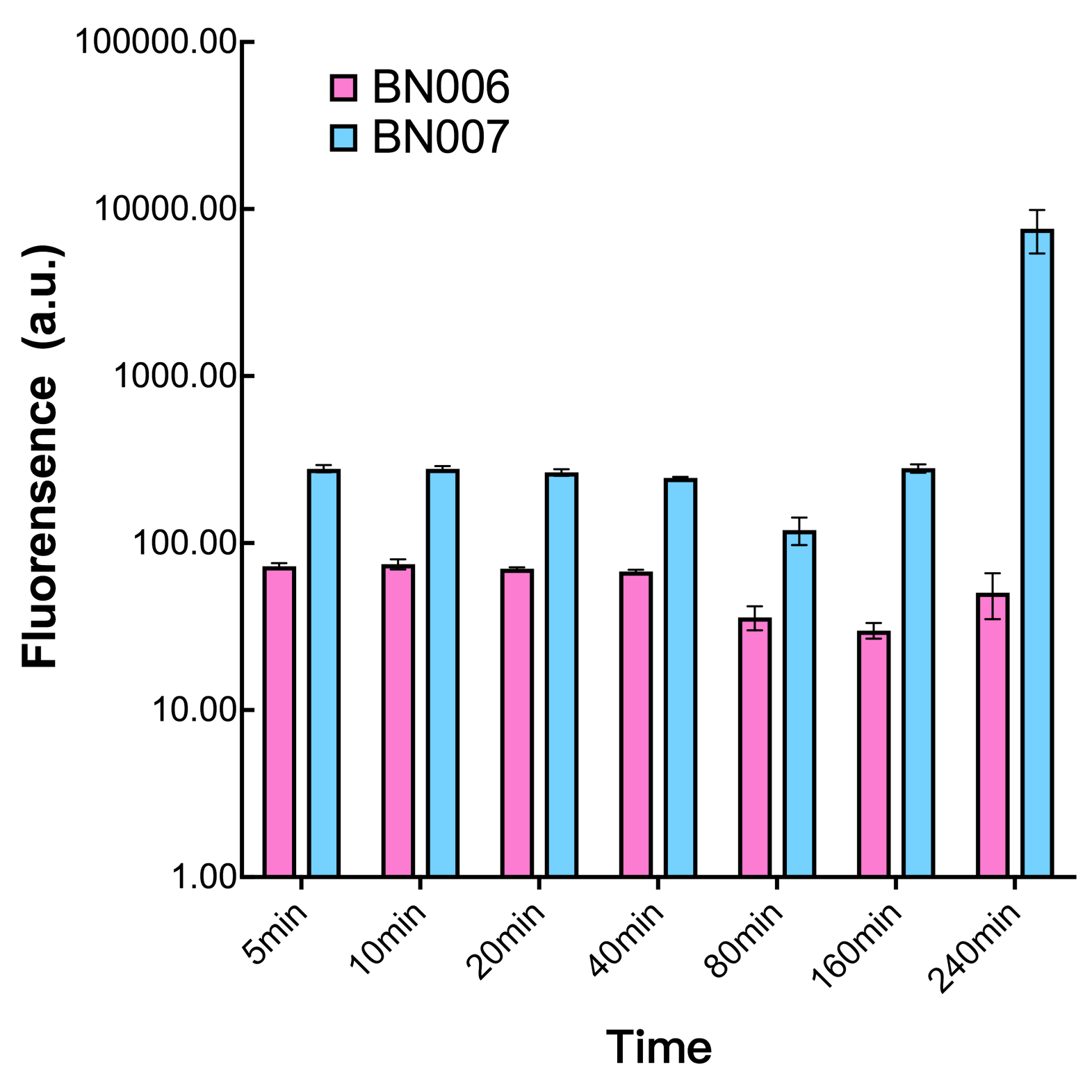
| + | [[File:T--BHSF_ND--Leakage experiment 3.png|429px|thumb|left|alt=Comparison between BN006/Contains BBa_K3202029 and BN007/Contains BBa_K3202030|Figure3: Time induction curve pBAD(BN007/Contains BBa_K3202030) at the concentration of 1mM compared to backbone (BN006/Contains BBa_K3202029)]] | ||
| + | |||
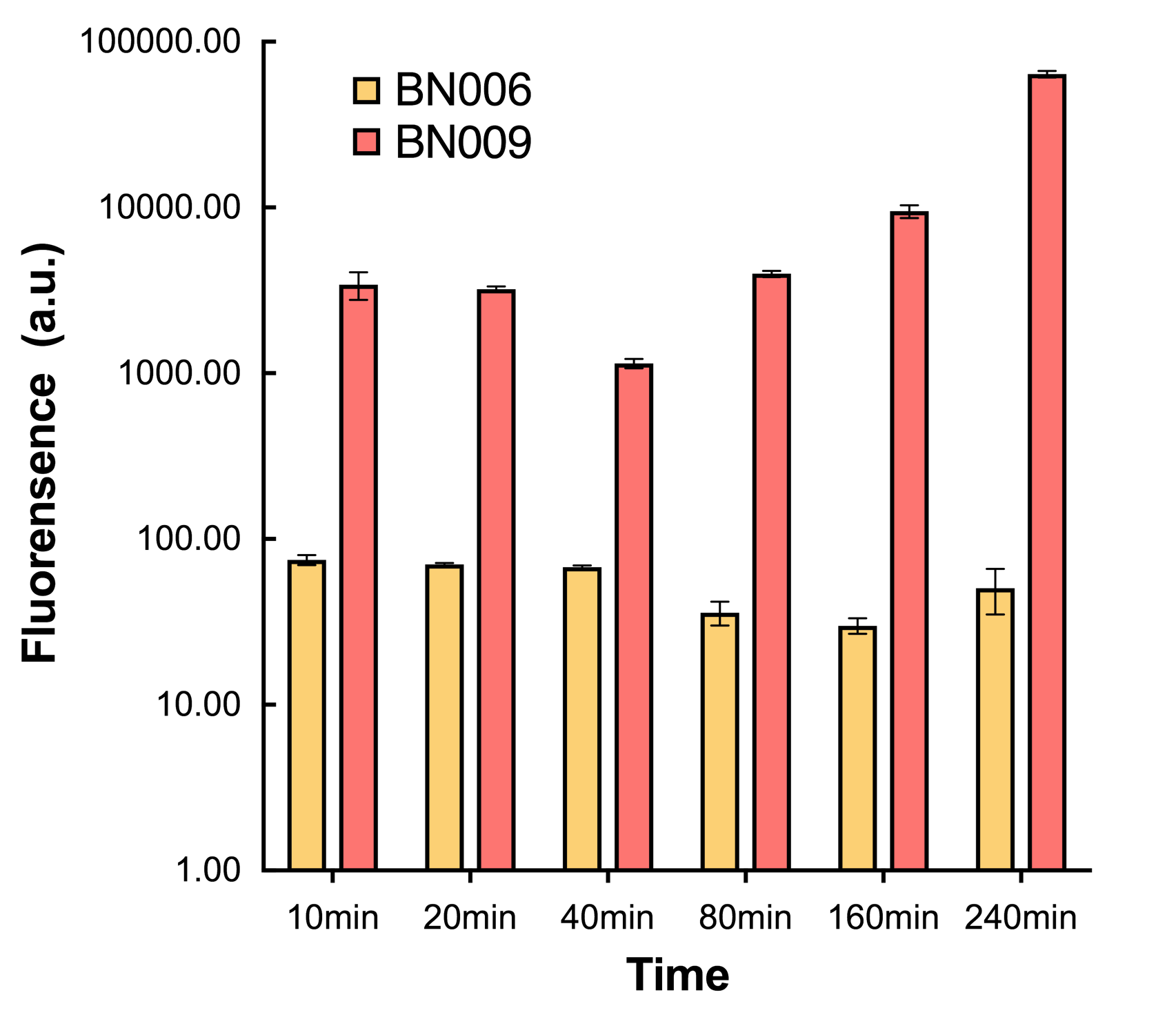
| + | [[File:T--BHSF_ND--Leakage experiment 4.png|435px|thumb|right|alt=Comparison between BN006/Contains BBa_K3202029 and BN009/Contains BBa_K3202031|Figure4: Time induction curve for Xyls(BN009/Contains BBa_K3202031) at the concentration of 1mM compared to backbone (BN006/Contains BBa_K3202029)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | === III.Experiment & Results === | ||
| + | |||
| + | In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication. | ||
| + | |||
| + | Two inducers with their respective promoters (AraC & pBAD; Xyls & Pm) are coupled with sfGFP to see if there is actually expression leakage when inducer is present quantitatively through flow cytometry. We tested fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration. Theoretically when inducers (AraC and 3MBz ) are present, promoters (pBAD and Pm) are initiated therefore both sfGFP are expressed; while sfGFP shouldn’t be expressed if inducer is absent. | ||
| + | When measured at the end of 240 minutes without inducer, BN009/Contains BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/Contains BBa_K3202029(backbone without GFP). BN007/Contains BBa_K3202030(pBAD+sfGFP) also showed a leakage of 90 a.u. The result suggested that both system have noticeable leakage and Xyls has much higher leakage. | ||
| + | |||
| + | When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/Contains BBa_K3202029 stays at a constant level, whereas the fluorescence of both Xyls and pBAD have increased exponentially. This is another sign that both system function properly. | ||
| + | |||
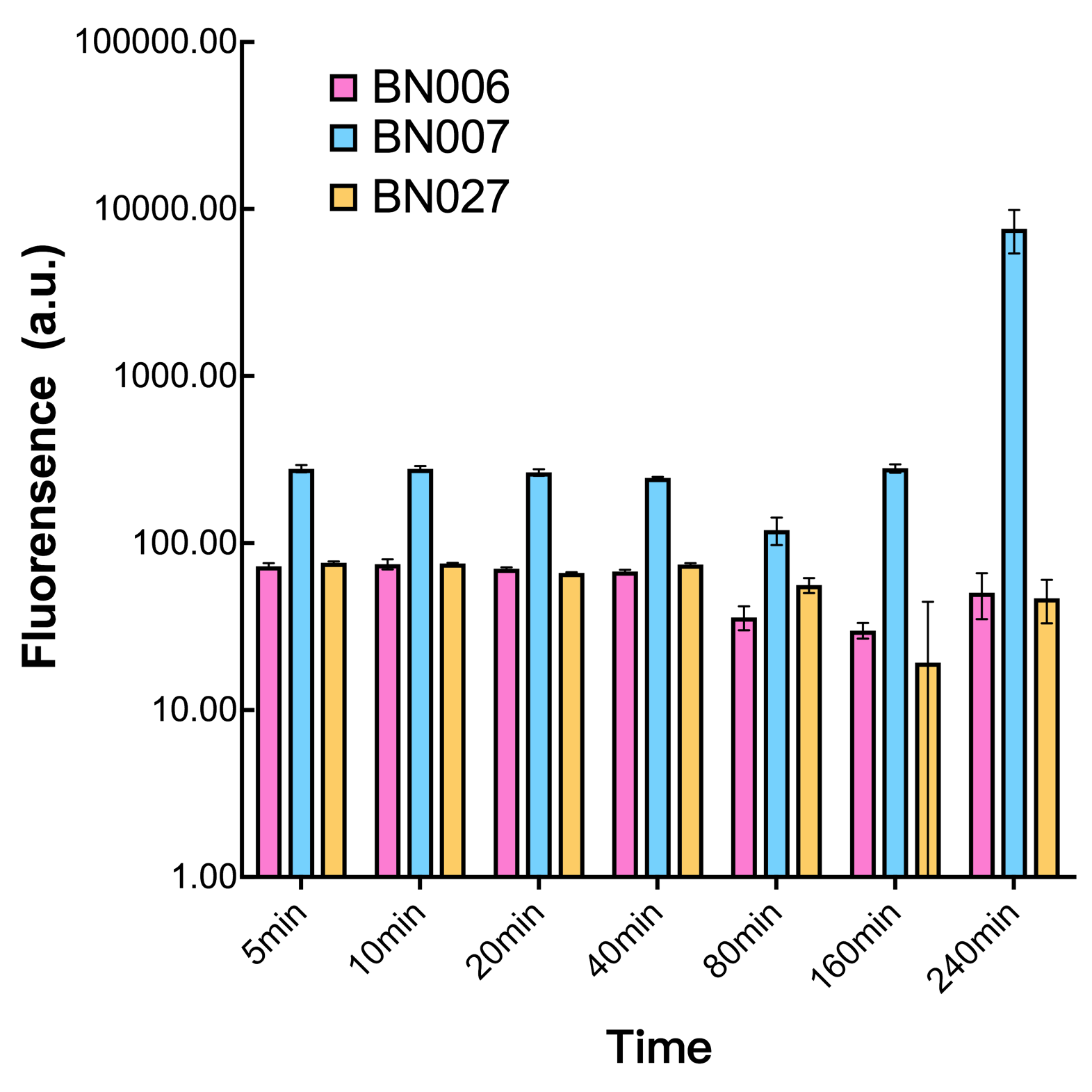
| + | We found an intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/Contains BBa_K3202030 is 200 a.u. higher than BN006/Contains BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone. Same thing happened to BN009/Contains BBa_K3202031. Therefore, in the subsequent experiments, systems are induced to up to 4 hours to ensure the accuracy of the experiment. Results shown above verified the presence of expression leakage of both systems when inducer is not expressed through the detectable fluorescence of reporter protein using flow cytometry. | ||
| + | |||
| + | |||
| + | =<b>Bistable System Module</b>= | ||
| + | |||
| + | <hr style="height:0px;border:none;border-top:none solid #555555;" /> | ||
| + | |||
| + | ===I.Background=== | ||
| + | |||
| + | We are inspired by a lately published article written by Belén Calles, Angel Goñi-Moreno, and Víctor de Lorenzo to design a double-bistable system which achieves zero basal expression of gene of interest in the absence of any inducer while ensuring constant transcriptional capacities and induction rates among promoters. | ||
| + | |||
| + | “The mechanism of a bistable system is elaborated below: a repressor transcriptionally inhibits the promoter which is responsible for the transcription of sRNA, while the sRNA translationally inhibits the repressor. The gene of interest is translationally coupled to the repressor gene. That is to say, the repressor protein R targets the strong promoter which the inhibitory sRNA is produced, so that R and sRNA are mutually inhibitory. Such arrangement helps to minimize the basal activity of the inducible module and thus suppress leaky gene expression levels.” | ||
| + | |||
| + | ===II.Design=== | ||
| + | |||
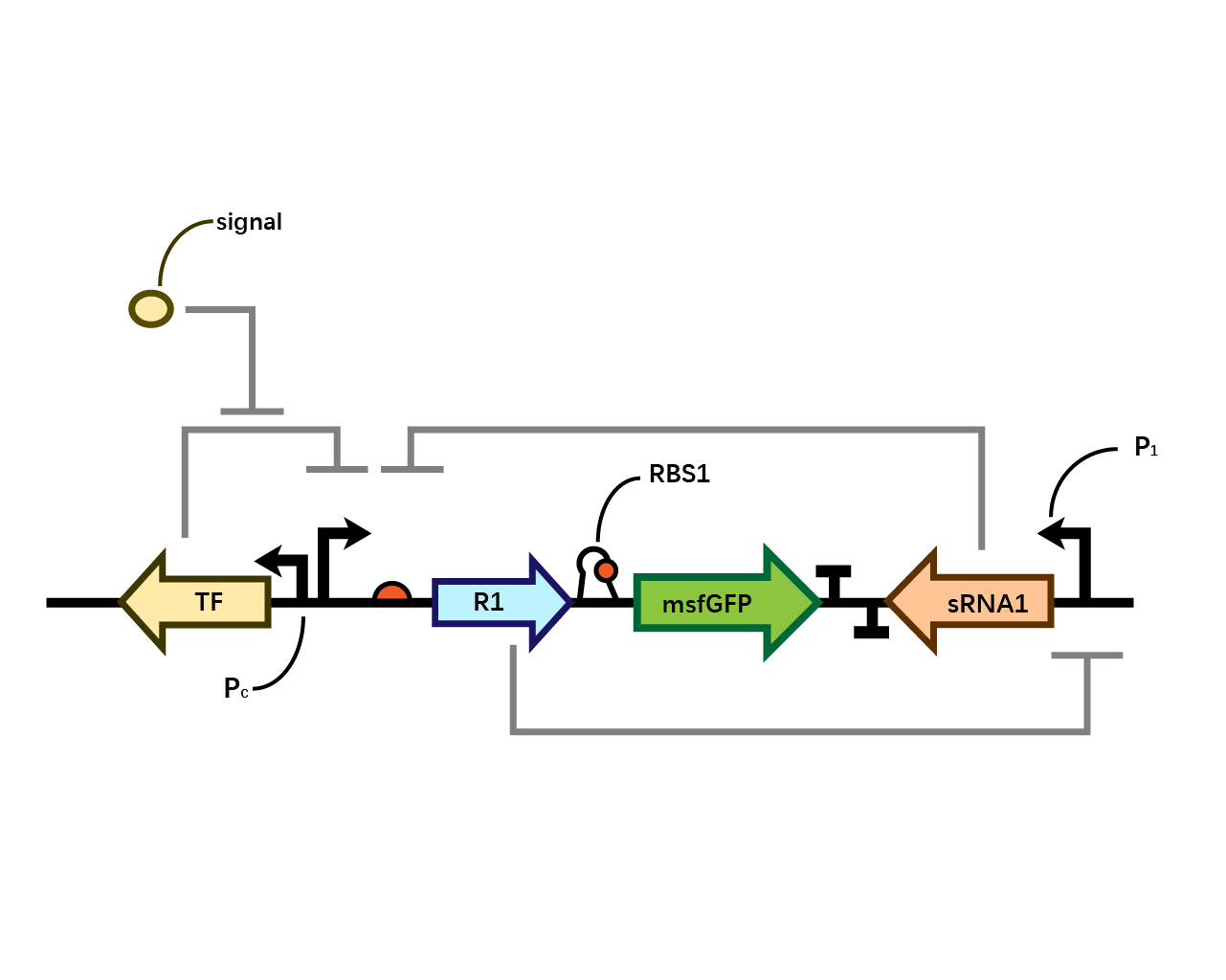
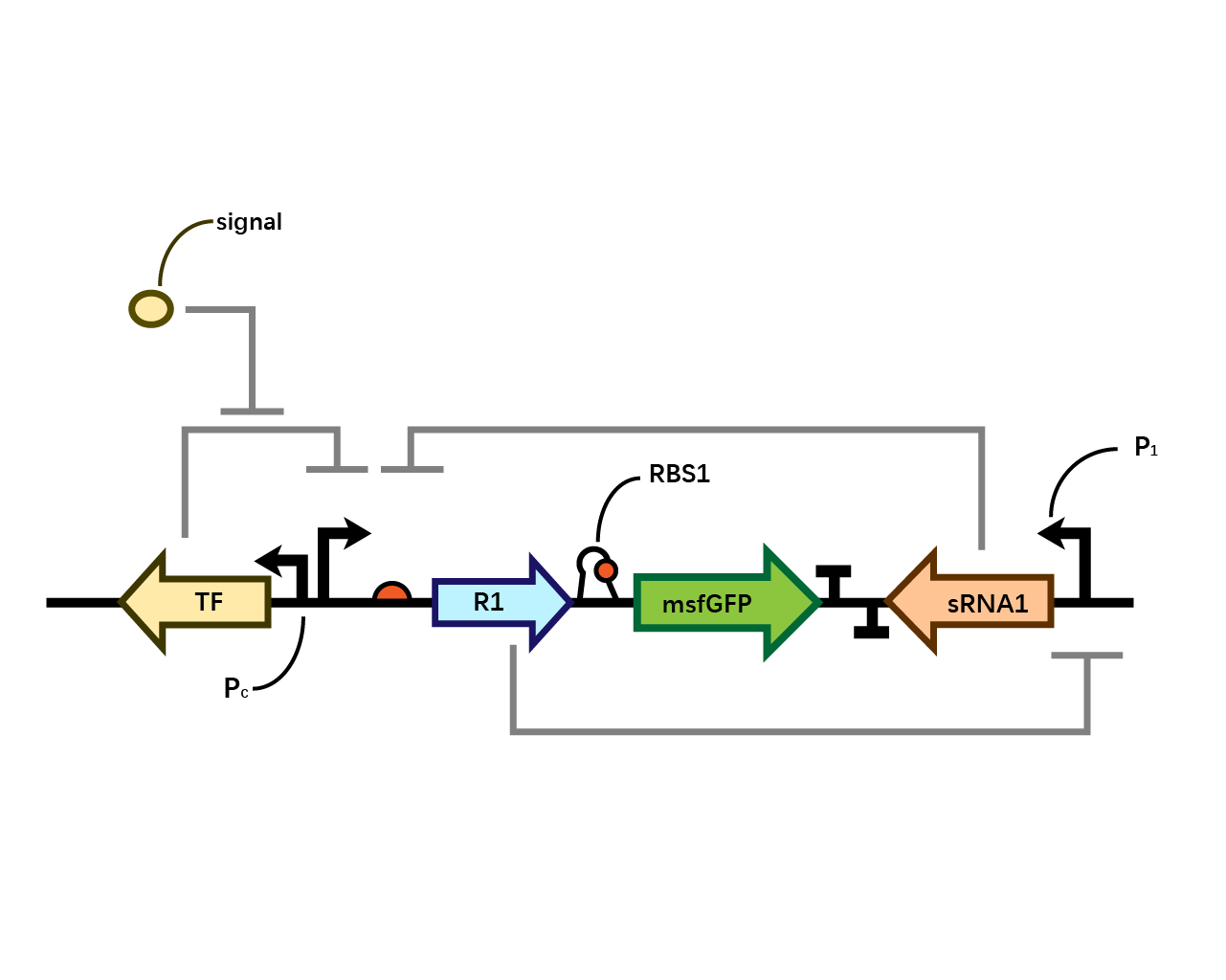
| + | In our system(Fig. 1), we have transcriptional repressor R1 expressed through the inducible promoter but translationally inhibited by MicC sRNA1 since the RBS of R1 is directly inhibited by sRNA while they initiates the transcription of the two repressors; the gene of interest is translationally coupled to the repressor gene while three promoters which produces the inhibitory sRNA are targeted by their corresponding repressors. Therefore, repressors and sRNAs are mutually inhibitory. | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable design 1.png |550px|thumb|center|alt=Basic Design of Bistable System |Figure 1]] | ||
| + | |||
| + | ====RBS1==== | ||
| + | |||
| + | The control of the recombinase in coordination with the upstream repressor gene, which is the target of the sRNA, requires a translational coupler cassette. “An efficient mechanism to reach this goal is based on the ability of translating ribosomes to unfold mRNA secondary structures. In our design, translational coupling is achieved by occluding the RBS of gene of interest by formation of a secondary mRNA structure, containing a His-tag sequence added to the 3 ́end of the repressor gene. The hairpin structure prevents the ribosomal recruitment to the recombinase and therefore its translation. In contrast, when the upstream repressor mRNA is actively translated, the 70s ribosome disrupts inhibitory mRNA secondary structure in the downstream gene translation initiation region thus allowing its expression.” | ||
| + | |||
| + | However, during the experiment we tested that the RBS1 (Fig. 2) sequence provided by the original article fails to function in our circuit, we found RBS2 (Fig. 3) with a different working mechanism. The structure of AUGA in RBS2 acts as both the stop codon (AUG) of the former gene sequence, releasing the former peptide chain; takes a step (gene code) backward, pinpointing a new ribosome binding site, and initiated the translation of next gene sequence with the start codon (UGA). However, finally we still used RBS1 since the crux actually lied in another place. | ||
| + | [[File: T--BHSF ND--Bistable design 2.png |429px|thumb|left|alt=RBS1 from the Original Article |Figure 2]] | ||
| + | [[File: T--BHSF ND--Bistable design 3.png |429px|thumb|right|alt=RBS1 with a Different Working Mechanism |Figure 3]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ====sRNA==== | ||
| + | |||
| + | Small non‐coding RNAs (sRNAs) have important functions as genetic regulators in prokaryotes. “sRNAs act post‐transcriptionally through complementary pairing with target mRNAs to regulate protein expression. Antisense sRNAs negatively regulates proteins by destabilizing the target protein's mRNA. Antisense sRNAs prevent translation by binding to the target mRNAs in a process mediated by the RNA chaperone Hfq.” On binding, both the mRNAs and sRNAs are degraded, suggesting that prokaryotic sRNAs—unlike their eukaryotic counterparts—act stoichiometrically on their targets. | ||
| + | |||
| + | ====Repressor==== | ||
| + | |||
| + | “A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA.” | ||
| + | |||
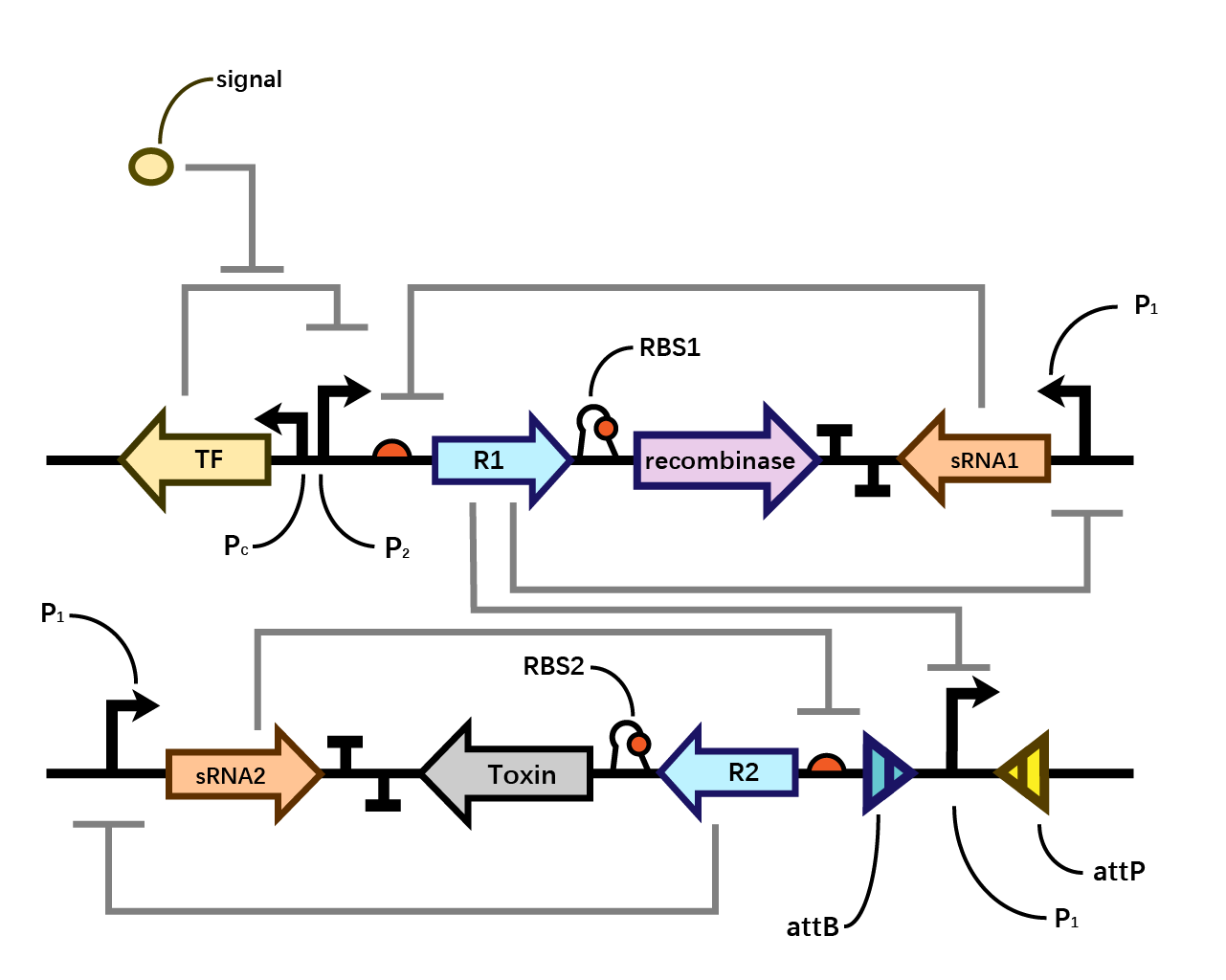
| + | We assume that when the whole system is applied to a factory, the situation where inducer is present is in the working condition of the bacterium. Within one single layer of the bistable system, the transcriptional factor is turned on, Pm initiates the expression of R1, which raises the concentration of R1 and thereby exerts greater inhibition on PR1 and therefore decreases the concentration of sRNA1. As sRNA1 is inhibited the inhibitory effect it exerts on R1 accordingly decreases, therefore the recombinase is expressed through the first layer, and vice versa. Nevertheless, given that the expression of R1 inhibits PR1, even if the recombinase is expressed PR1 can be flipped over but cannot express the second layer. Since there is no further input signal to the second layer of the bistable system, the inhibitory effect of sRNA2 dominates over R2, therefore, both R2 and the toxin cannot be expressed. That is to say, the additional second layer ensures that there is no leakage of toxin when inducer is present. | ||
| + | |||
| + | However, when the bacterium is stolen and thereby inducer isn’t present, the transcriptional factor is turned off, promoter Pm doesn’t work. Therefore, sRNA1 dominates over R1 and then the recombinase is not expressed in the first layer. Since there is a decrease in the expression of R1, the inhibitory effect it exerts on PR1 decreases, therefore PR1 which has already been flipped over when inducer is present (working condition) initiates the expression of R2. Applying the same logic, finally R2 dominates over sRNA2 and expresses the toxin to kill the cell. (Fig. 4) | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable design 4.png |550px|thumb|center|alt=Complete Version of Our Design |Figure 4]] | ||
| + | |||
| + | ====Why double layer?==== | ||
| + | |||
| + | The reason why we designed such double-bistable system can be attributed to the consideration that the double-decked bistable systems of both recombinase and toxin could further reinforce the insurance of zero expression of toxin. | ||
| + | |||
| + | When the second bistable system coupling the toxin with the first layer of recombinase, double insurance of zero expression of recombinase and toxin is achieved. However, as the diagram shown below, a single bistable system where toxin is coupled without the second layer of insurance could collapse if the mutual inhibitory effect of repressor and sRNA within the first layer malfunctions. Whereas such concern could be eliminated by the second layer of toxin with a bistable system around. | ||
| + | |||
| + | ===III. Experiment & Results=== | ||
| + | |||
| + | After the circuit (Fig. 5) including only one layer of the bistable system is constructed with recombinase being replaced by sfGFP, LacI & TetR serving as repressor, and used 3MBz & AraC as inducers, we qualitatively tested the fluorescence of the reporter protein under two situations when excited blue light. Theoretically, sfGFP should be expressed when inducer is present since LacI, which transcribes sfGFP, dominates over the inhibitory force of sRNA. However, in the experiments when inducers are present and absent, there is no fluorescence of sfGFP (BN027/Contains BBa_K3202074 & 28/Contains BBa_K3202075 & 29/Contains BBa_K3202076 & 30/Contains BBa_K3202077). We deduced that there must be defects lying in our circuit. | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable Experiment 1.png |550px|thumb|center|alt=Single Layer of Bistable System |Figure 5]] | ||
| + | |||
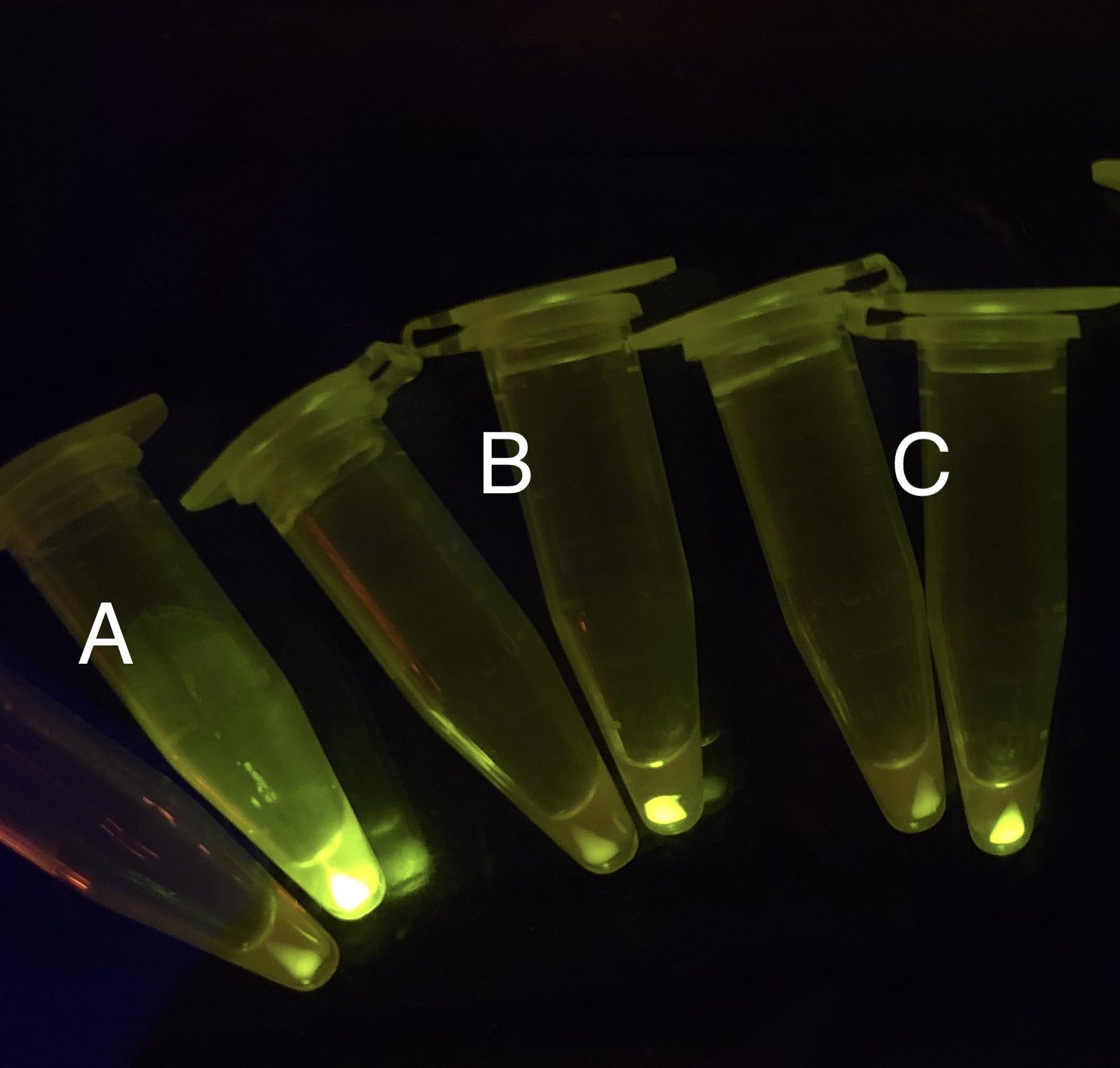
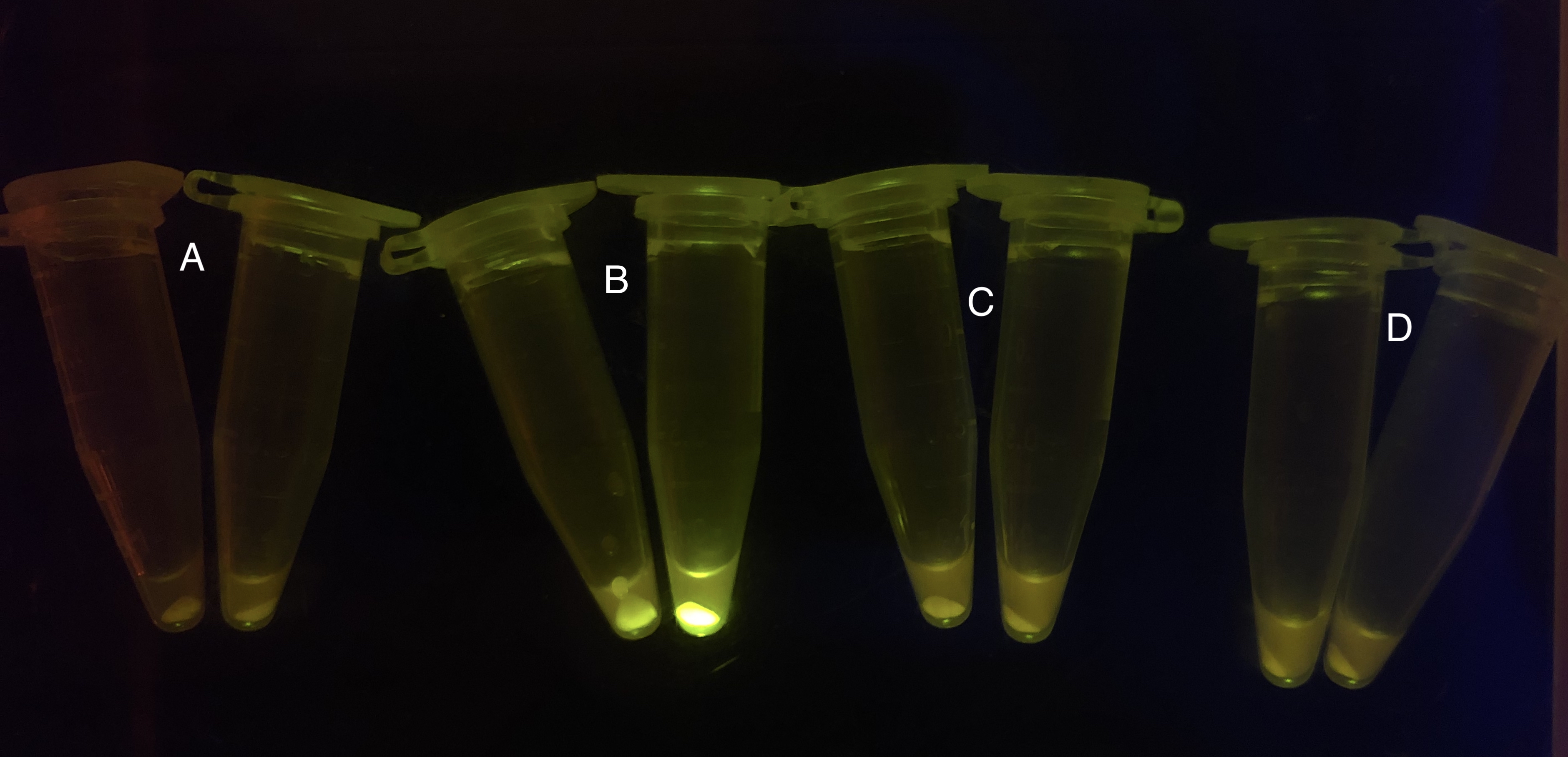
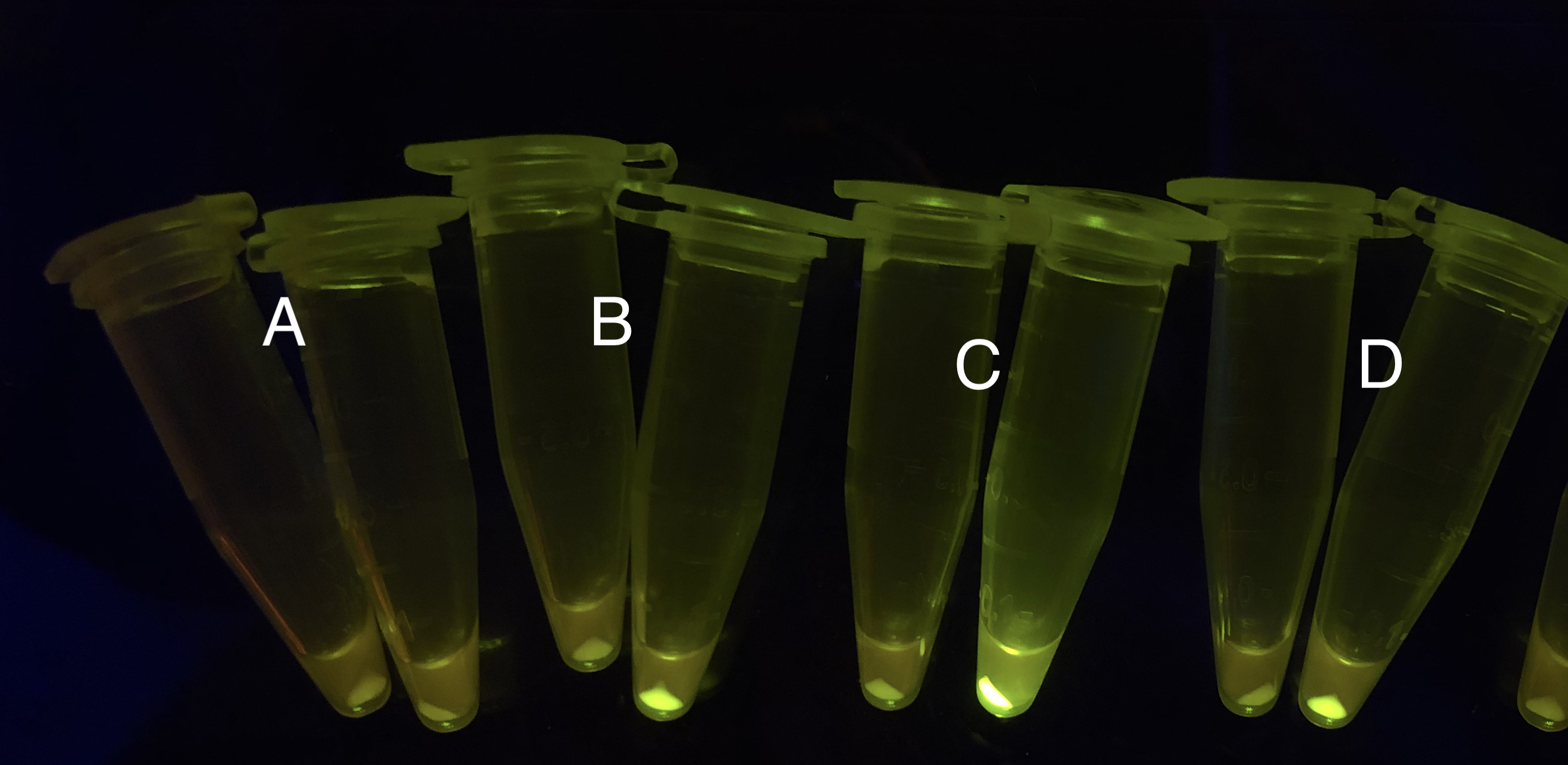
| + | [[File: T--BHSF ND--Bistable experiment 2.png |440px|thumb|left|alt= Qualitative Comparison between BBa_K3202029 & BBa_K3202030 & BBa_K3202074 & BBa_K3202076 |Figure 6 A: BN006/Contains BBa_K3202029 (without GFP) when inducer is absent and present; B: BN007/Contains BBa_K3202030 (AraC + sfGFP) when inducer is absent and present; C: BN027/Contains BBa_K3202074 (AraC + LacI + sfGFP) when inducer is absent and present; D: BN029/Contains BBa_K3202076 (AraC + TetR + sfGFP) when inducer is absent and present]] | ||
| + | |||
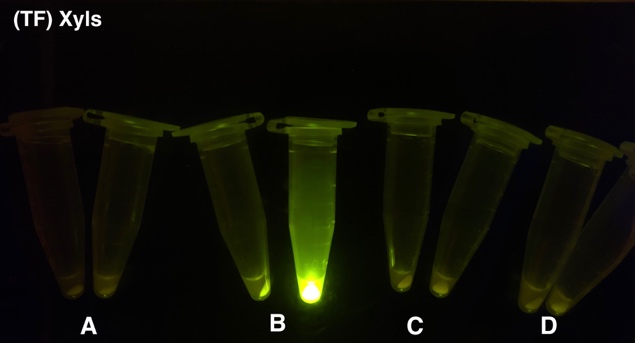
| + | [[File: T--BHSF ND--Bistable Experiment 2.png |400px|thumb|right|alt=Qualitative Comparison between BBa_K3202029 & BBa_K3202030 & BBa_K3202075 & BBa_K3202077 |Figure 7 A: BN006/Contains BBa_K3202029 (without GFP) when inducer is absent and present; B: BN007/Contains BBa_K3202030 (Xyls + pBAD + sfGFP) when inducer is absent and present; C: BN028/Contains BBa_K3202075 (Xyls + LacI + sfGFP) when inducer is absent and present; D: BN030/Contains BBa_K3202077 (Xyls + TetR + sfGFP) when inducer is absent and present]] | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable Experiment 4.png |429px|thumb|left|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202030 & BBa_K3202074 |Figure 8: As shown in the graph from flow cytometry experiments: in the presence of a fixed concentration of inducer (1.0 mM of Ara) at the indicated time points (5 to 240 min after induction), BN006/Contains BBa_K3202029, which carries no sfGFP, has low fluorescence level under 100 a.u. being expressed in inducer’s presence. BN027/Contains BBa_K3202074 (under the control of bistable system) shows similar pattern as time increases as that of BN006/Contains BBa_K3202029. BN007/Contains BBa_K3202030 (without control of bistable system) showed higher fluorescence level throughout the experiment]] | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable Experiment 5.png |429px|thumb|right|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202030 & BBa_K3202074 |Figure 9: As shown in the graph from flow cytometry experiments: at same time period, as the conc. of Ara increases from 0 to 1.0 mMol, BN006/Contains BBa_K3202029 (no sfGFP) and BN027/Contains BBa_K3202074 (under the control of bistable system) showed both fluorescence level around 100 a.u., revealing a lack of expression of sfGFP; while BN007/Contains BBa_K3202030 displayed increase in fluorescence level as inducer concentration increases]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Analyzing through the whole circuit, we deduced that the crux might lie in RBS1 for the sake of its overwhelmingly strong hair-pin structure that even if the 3’ end of LacI is translated the ribosomes produced still couldn’t melt sRNA secondary structure, thereby sfGFP cannot be translated. Therefore, we replaced RBS1 with a commonly used RBS: B0034 with weaker strength in order to ensure that there is no problem lying in other sites in the circuit. We tested the sfGFP’s fluorescence in inducers’ presence under excitation of blue light. The expression of sfGFP in the second attempt (Fig. 10B, Fig. 11B & D) proved to us that there’s nothing wrong with the rest of the circuit except for RBS1. | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable experiment 6.jpeg |395px|thumb|left|alt=Qualitative Comparison between BBa_K3202047 & BBa_K3202031 |Figure 10 A: BN040/Contains BBa_K3202047 (Xyls + TetR + RBS B0034 + sfGFP) when inducer is present and absent; B: BN009/Contains BBa_K3202031 (Xyls + msfGFP) when inducer is present and absent]] | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable experiment 7.jpeg |438px|thumb|right|alt=Qualitative Comparison between BBa_K3202030 & BBa_K3202046|Figure 11 A: BN007/Contains BBa_K3202030 (AraC + pBAD + sfGFP) when inducer is absent; C: BN007/Contains BBa_K3202030 (AraC + pBAD + sfGFP) when inducer is present; B: BN036/Contains BBa_K3202046 (pBAD + LacI +RBS B0034 + sfGFP) when inducer is absent; D. BN036/Contains BBa_K3202046 (pBAD + LacI +RBS B0034) + sfGFP when inducer is present]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
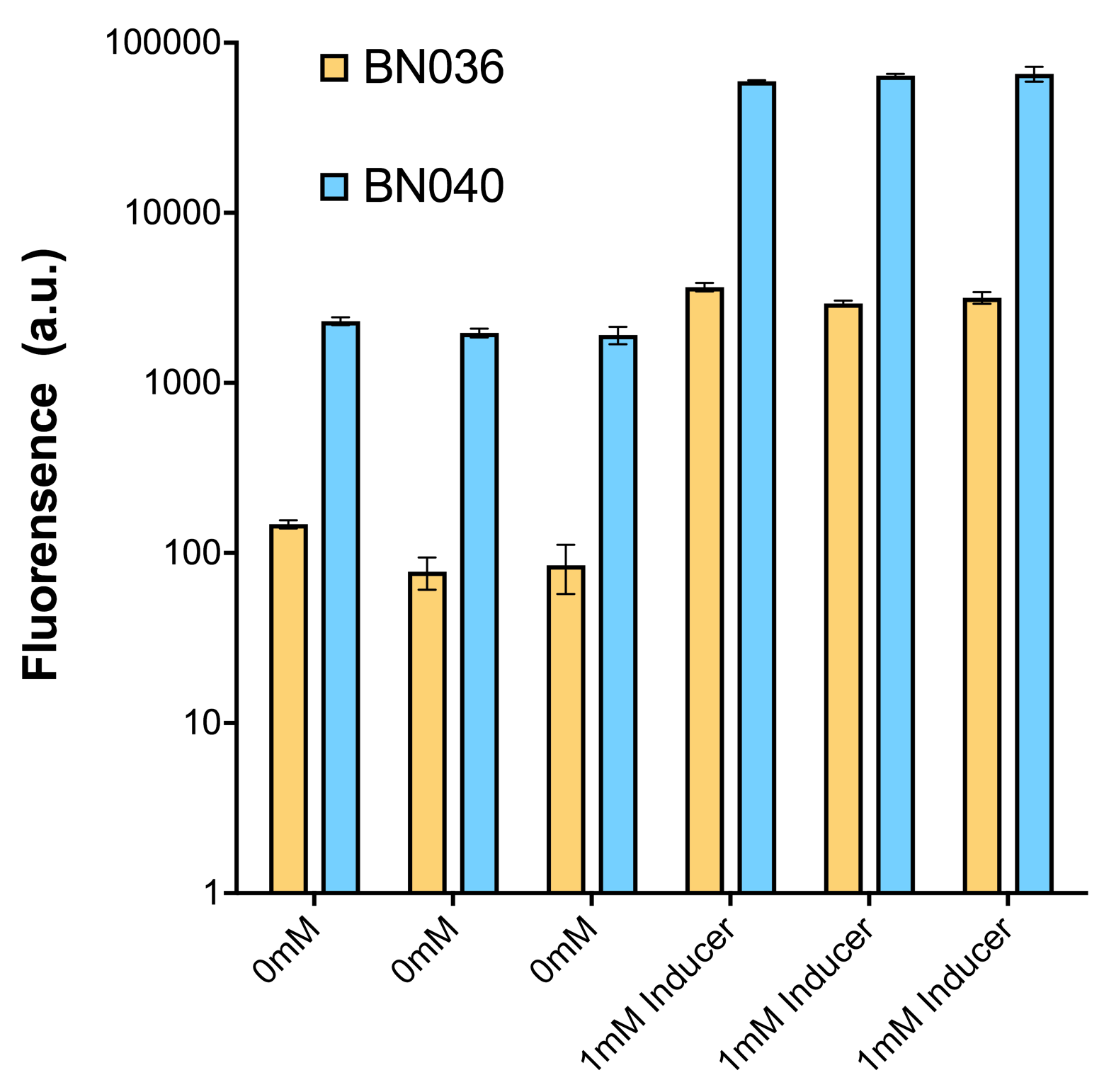
| + | [[File: T--BHSF ND--Bistable Experiment 8.png |350px|thumb|center|alt=Quantitative Comparison between BBa_K3202046 & BBa_K3202047|Figure 12: As shown in the graph from flow cytometry experiments: at same time period, in the absence and presence(1mMol) of inducers, both plasmids (BN036/Contains BBa_K3202046 & BN040/Contains BBa_K3202047) have their fluorescence level 10 times lower in inducer’s absence than that in inducer’s presence]] | ||
| + | |||
| + | |||
| + | Through further searching we found that RBS2 has a weaker hair pin structure while it could support the translation of repressor. We replaced RBS B0034 with RBS2. Results are shown in the two graphs below. Plasmid with RBS2 expressed sfGFP with quite a high expression leakage when both inducers are absent. We thought that such high expression leakage might be the result of the weak hair-pin structure of RBS2, which means that we still need a strong RBS to reduce expression leakage. Hence, by we were inspired to realize that we might wrongly include the terminator of LacI before RBS. If we include the terminator, the transcription of LacI stops there and therefore the LacI protein produced by translation ends in front of RBS and leaves the mRNA secondary structure before melting it, sfGFP failed to be expressed. | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable experiment 9.jpeg|420px|thumb|left|alt=Qualitative Comparison between BBa_K3202031 & BBa_K3202049 & BBa_K3202050 |Figure 13 A:BN009/Contains BBa_K3202031 (Xyls + sfGFP) when inducer is absent and present; B:BN045/Contains BBa_K3202049 (TetR + Xyls +RBS2) when inducer is absent and present; C:BN049/Contains BBa_K3202050 (LacI+ Xyls + RBS2) when inducer is absent and present]] | ||
| + | |||
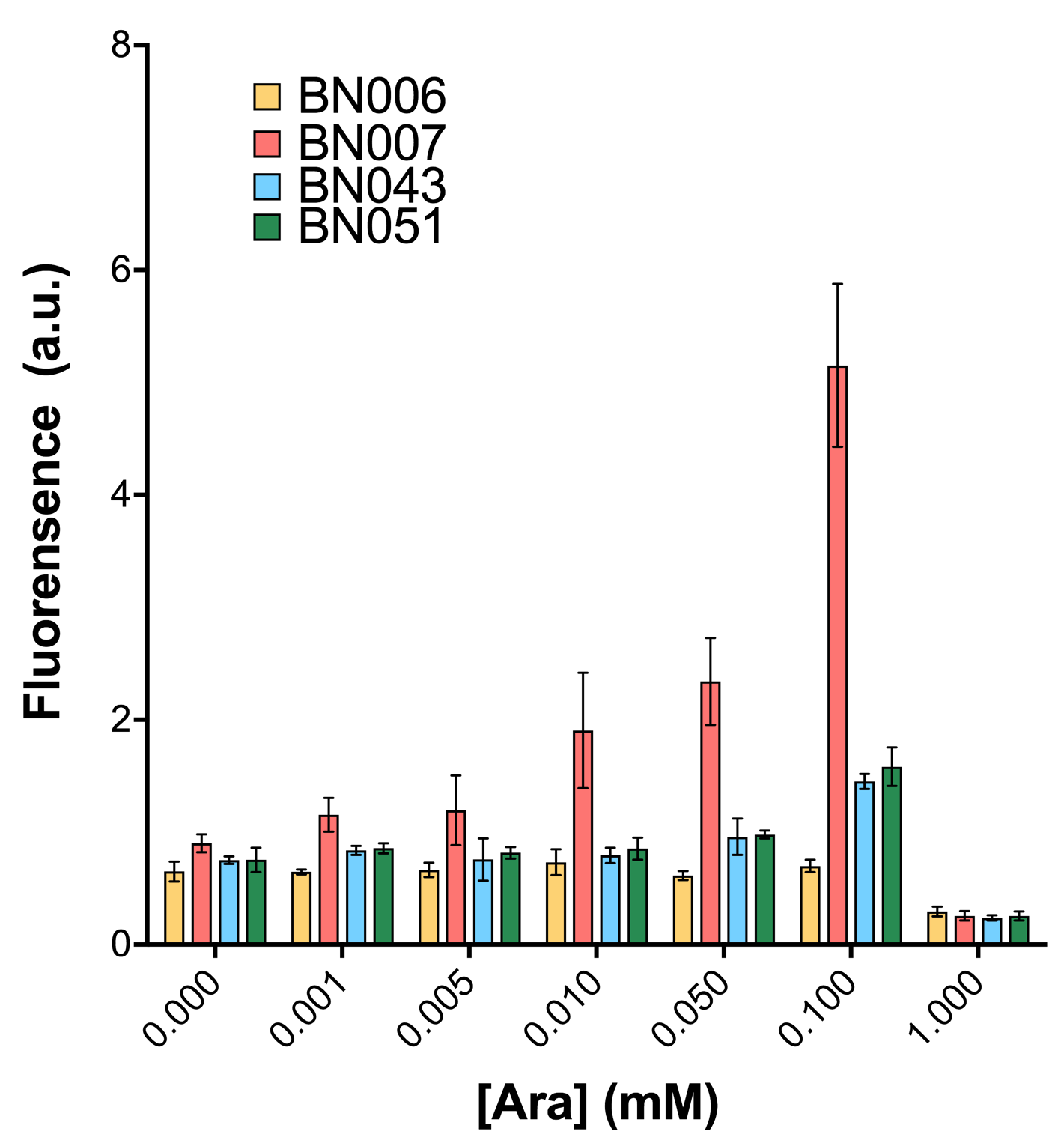
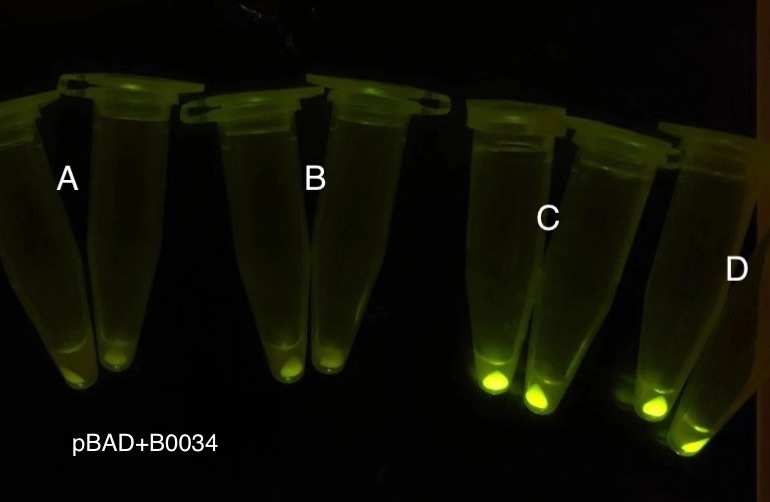
| + | [[File: T--BHSF ND--Bistable experiment 10.png|440px|thumb|right|alt=Qualitative Comparison between BBa_K3202029 & BBa_K3202048 & BBa_K3202030 & BBa_K3202051|Figure 14 A:BN006/Contains BBa_K3202029 (without GFP) when inducer is absent and present ; B:BN043/Contains BBa_K3202048 (pBAD + LacI +RBS2 + sfGFP) when inducer is absent and present; C:BN007/Contains BBa_K3202030 (AraC + pBAD + sfGFP) when inducer is absent and present; D:BN051/Contains BBa_K3202051 (pBAD + TetR + RBS2 + sfGFP) when inducer is absent and present]] | ||
| + | |||
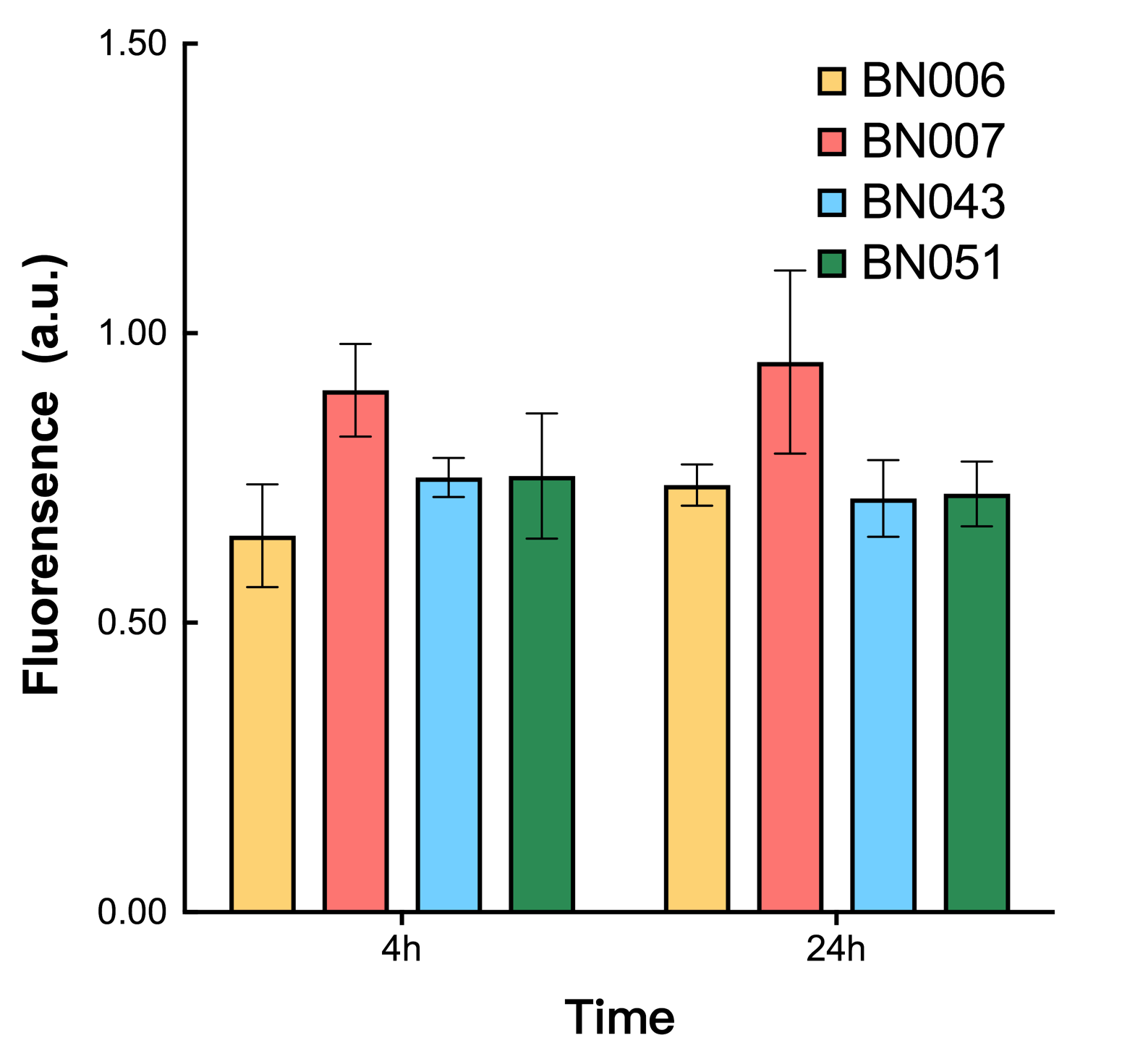
| + | [[File: T--BHSF ND--Bistable Experiment 11.png|420px|thumb|left|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202030 & BBa_K3202048 & BBa_K3202051|Figure 15]] | ||
| + | |||
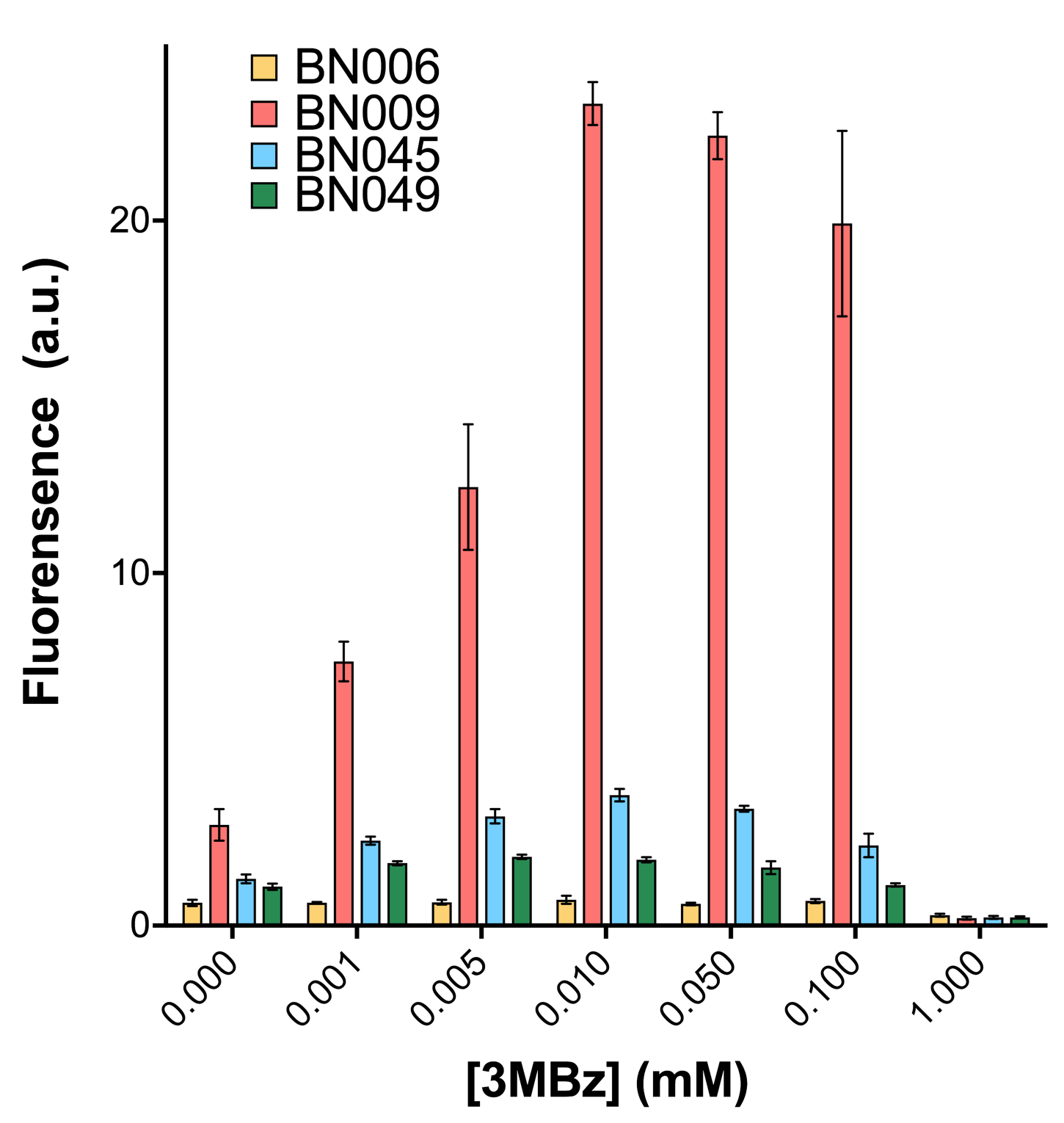
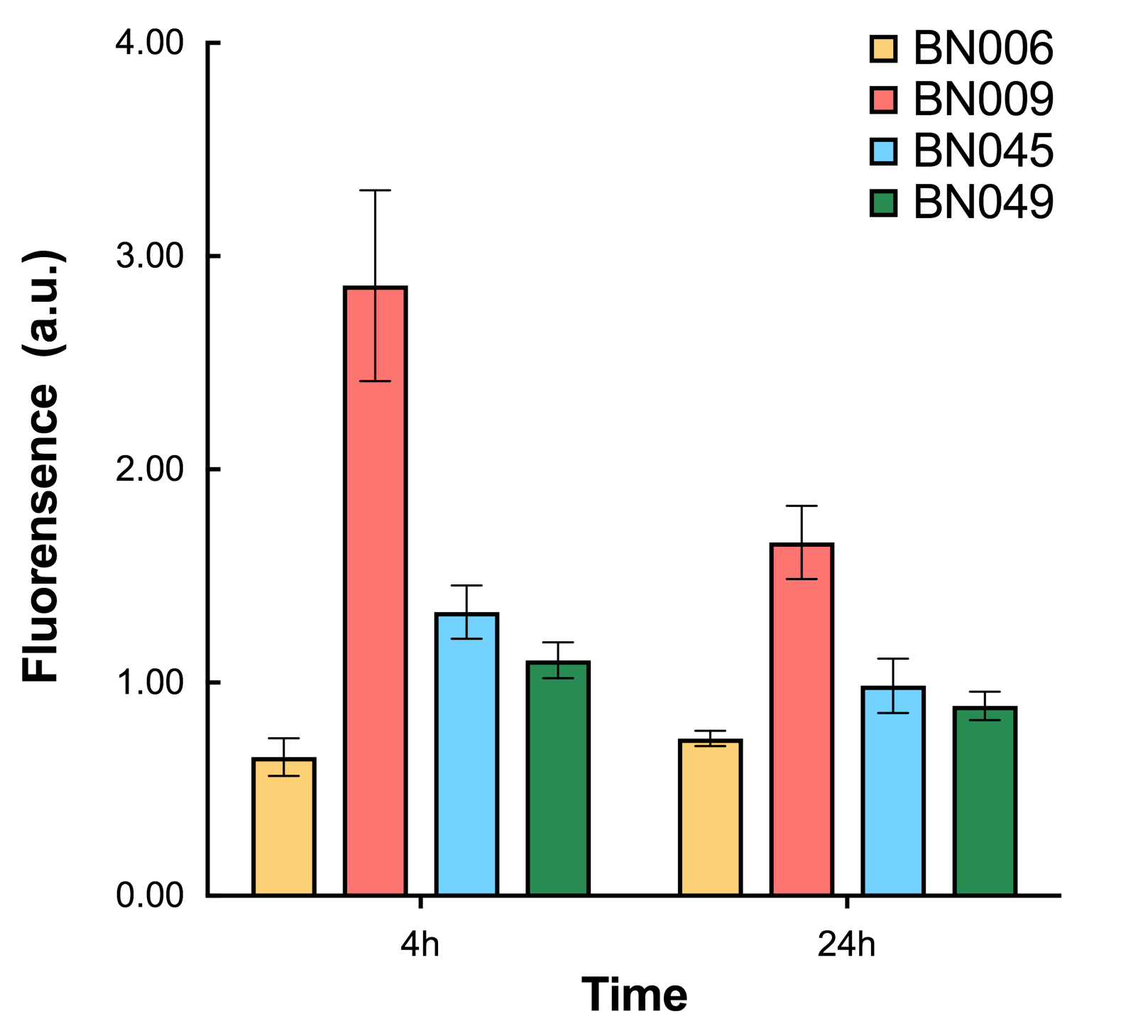
| + | [[File: T--BHSF ND--Bistable Experiment 12.png|430px|thumb|right|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202031 & BBa_K3202049 & BBa_K3202050 |Figure 16]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | As shown in Fig. 15 & 16 from flow cytometry experiments: at the same time period, the conc. of inducer increases from 0.000 to 1.000 mMol, BN006/Contains BBa_K3202029, which carries no sfGFP, has a low fluorescence level around 0.60 a.u. in inducer’s absence. BN007/Contains BBa_K3202030 and BN009/Contains BBa_K3202031 without the control of bistable system had their fluorescence value at a relatively higher level in inducer’s absence and their fluorescence value increase as conc. of inducers increases. BN043/Contains BBa_K3202048 & 51/Contains BBa_K3202051 & 45/Contains BBa_K3202049 & 49/Contains BBa_K3202050 (under the control of bistable system) have their fluorescence value lower than that of BN007/Contains BBa_K3202030 and BN009/Contains BBa_K3202031 in inducer’s absence and have their fluorescence value as conc. of inducer increases. | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable Experiment 13.png|428px|thumb|left|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202030 & BBa_K3202048 & BBa_K3202051|Figure 17]] | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable Experiment 14.png|428px|thumb|right|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202031 & BBa_K3202049 & BBa_K3202050 |Figure 18]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
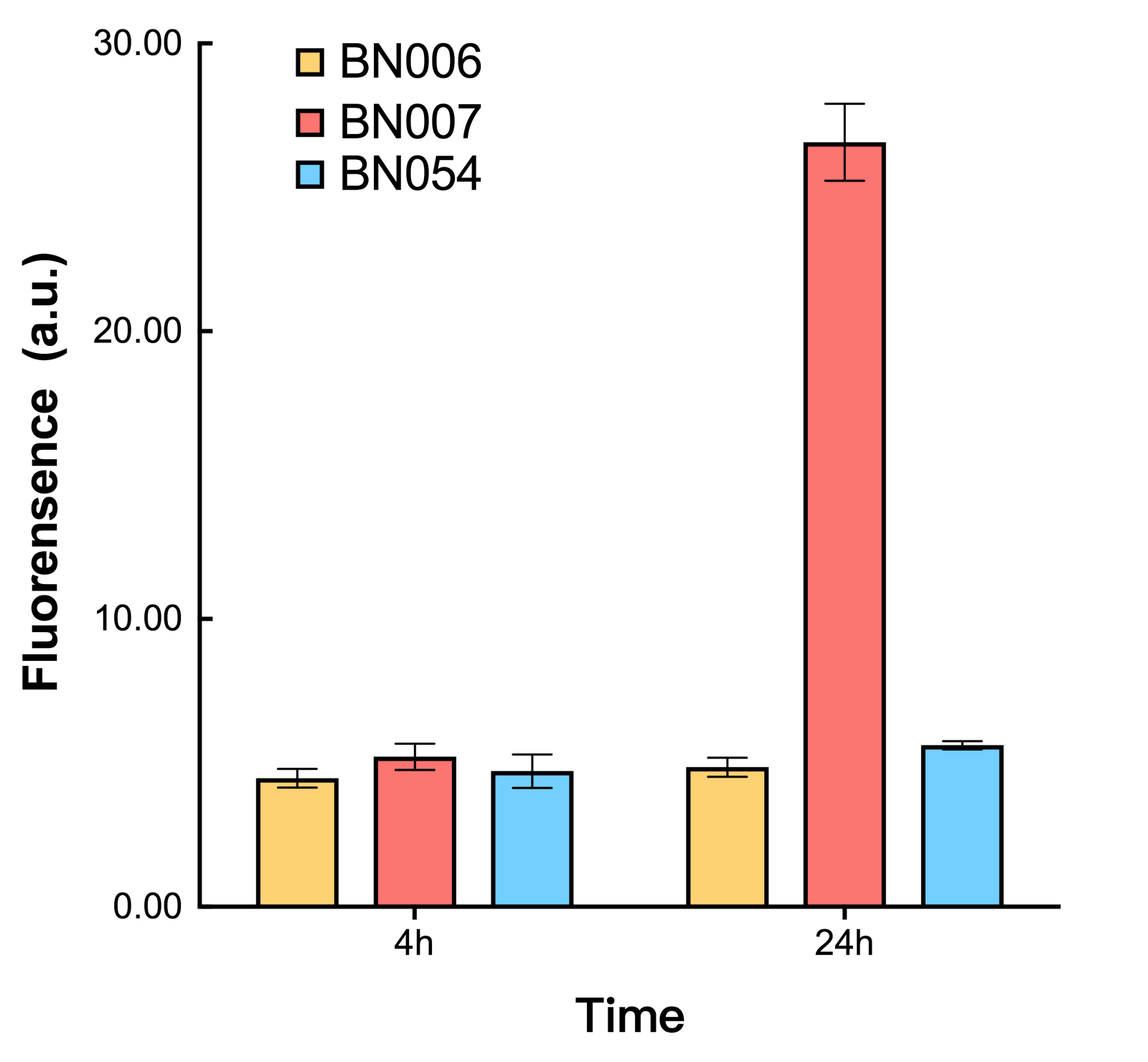
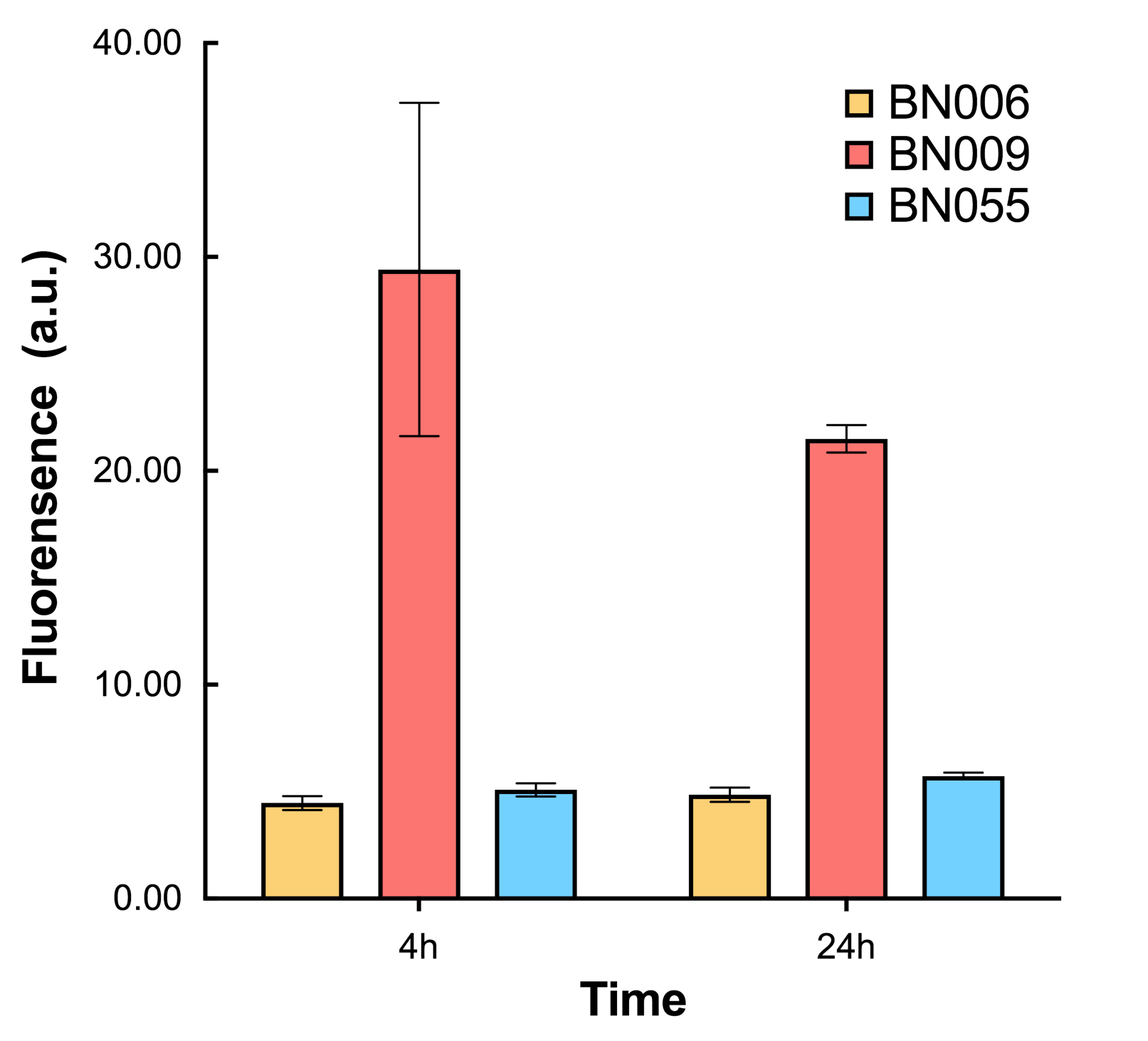
| + | As shown in Fig. 17 & 18 from flow cytometry experiments: in the absence of inducer at the indicated time points (4h and 24h after induction), BN006/Contains BBa_K3202029, which carries no sfGFP, has a low fluorescence level around 0.60 a.u. in inducer’s absence. BN043/Contains BBa_K3202048 & 51/Contains BBa_K3202051 & 45/Contains BBa_K3202049 & 49/Contains BBa_K3202050 (under the control of bistable system) have their fluorescence value higher than that of BN006/Contains BBa_K3202029 while lower than that of BN007/Contains BBa_K3202030 &BN009/Contains BBa_K3202031 (without the control of bistable system) in inducer’s absence. | ||
| + | |||
| + | We improved our gene circuit upon that reflection, with exclusion of the terminator of repressors and replacement of BRS2 using RBS1, and made the third trial. The results are shown below. | ||
| + | |||
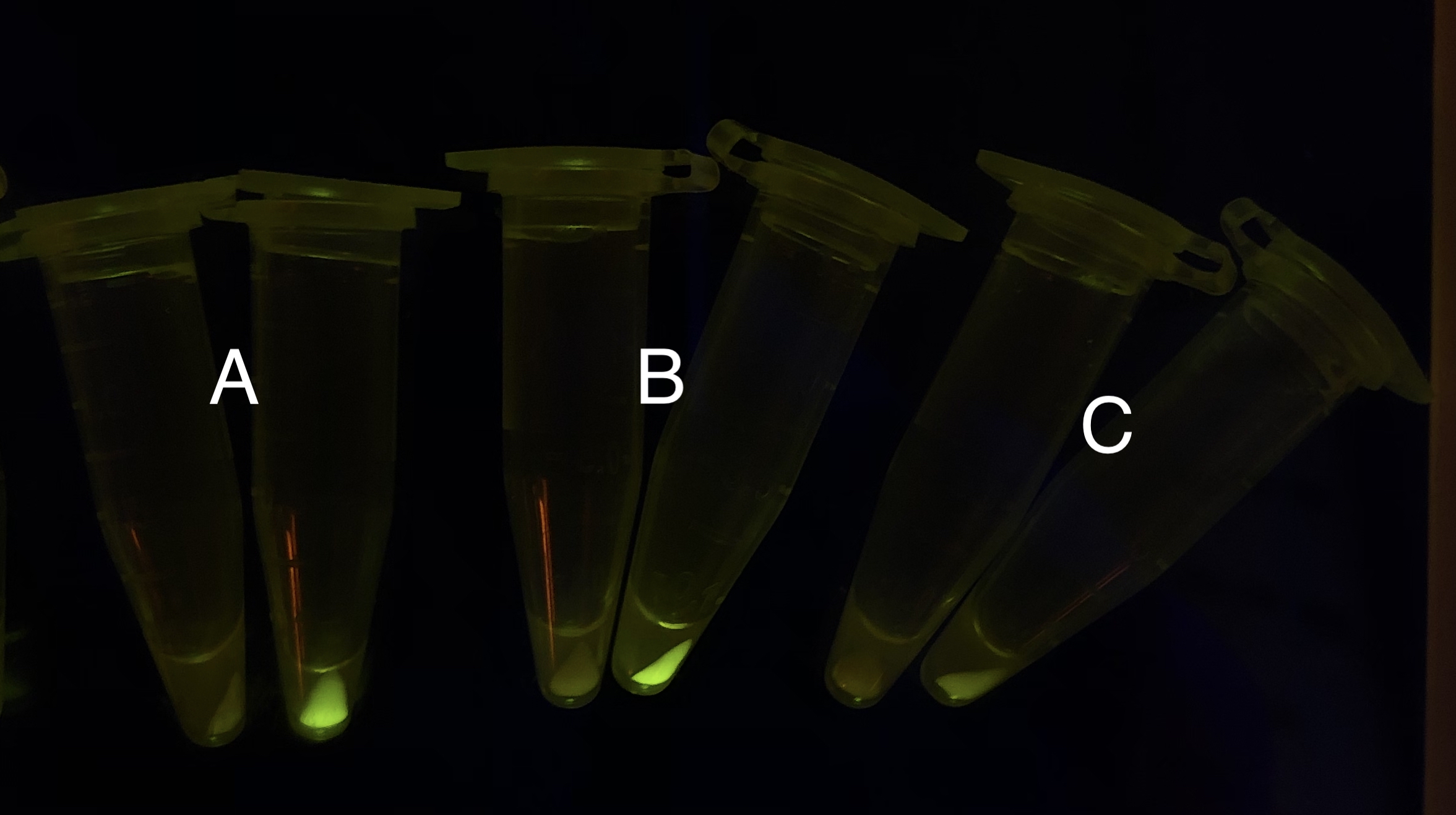
| + | [[File: T--BHSF ND--Bistable experiment 15.jpeg|428px|thumb|center|alt=Qualitative Comparison between BBa_K3202030 & BBa_K3202045 & Contains BBa_K3202046 |Figure 19 A: BN007/Contains BBa_K3202030 (AraC + pBAD + sfGFP) when inducer is absent and present; B: BN055/Contains BBa_K3202045 (Xyls + TetR + RBS1 + sfGFP) when inducer is absent and present; C: BN054/ Contains BBa_K3202044 (AraC + LacI +RBS1 + sfGFP) when inducer is absent and present]] | ||
| + | |||
| + | As shown in Fig. 20 & 21 from flow cytometry experiments: in the absence of inducer at the indicated time points (4h and 24h after induction), BN006/Contains BBa_K3202029, which carries no sfGFP, has a low fluorescence level around 0.60 a.u. in inducer’s absence. BN007/Contains BBa_K3202030 and BN009/Contains BBa_K3202031are two plasmids without the control of bistable system; they displayed high fluorescence level as time passed. BN054/Contains BBa_K3202044 and BN055/Contains BBa_K3202045 have similarly no fluorescence level as BN006/Contains BBa_K3202029. | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable Experiment 17.png|428px|thumb|left|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202030 & BBa_K3202044 |Figure 20]] | ||
| + | |||
| + | [[File: T--BHSF ND--Bistable Experiment 16.png|428px|thumb|right|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202031 & BBa_K3202045 |Figure 21]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| − | + | As shown in Fig. 22 & 23 from flow cytometry experiments: at same time period, as the conc. of inducers increases from 0.0001 (nearly no inducer) to 10 mMol, BN006/Contains BBa_K3202029, which carries no sfGFP, always has a low fluorescence level around 0.60 a.u. While both two plasmids (BN054/Contains BBa_K3202044, induced by Ara; BN055/Contains BBa_K3202045, induced by 3MBz) have their fluorescence level increase from around 10 a.u. (lack of expression of sfGFP) when inducer is absent to a higher value. | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | [[File:T-- | + | [[File: T--BHSF ND--Bistable Experiment 18.png|428px|thumb|left|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202044 |Figure 22]] |
| + | [[File: T--BHSF ND--Bistable Experiment 19.png|428px|thumb|right|alt=Quantitative Comparison between BBa_K3202029 & BBa_K3202045 |Figure 23]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
Latest revision as of 22:22, 20 October 2019
Pc-Xyls
This composite part is designed as a genetic circuit that is used as a control group of our later experiment.
This acts as a control group in our experiment. It is compared to BBa_K3202030 and BBa_K3202031 in the leakage/inducer module, and later used in the bistable system module.
Leakage/Inducer Module
I.Background
Gene expression depends largely on transcription and translation. Such processes display activated/non-activated states. For some situations, we would expect zero expression of GOI without the presence of inducer. However, leakage still exist which is due to the incomplete OFF stage of the transcriptional system. Even without proper inducers, GOI is still partly expressed in some case. Such leakage is called the basal leakage.In some cases, as long as one molecule of toxin protein, is translated, it can kill the entire cell. Thus, preventing leakage is a vital in regulating gene expression. Current methods of solving leakage problems includes introducing strong inducible promoters to the system. However, there are substantially no regulating system or transcriptional factor that can function as a technically ON/OFF switch to prevent basal level leakage. This may due to weak RNAP-DNA interactions or different transcriptional capacities and induction rate of different promoters and inducers.
II.Design
We mainly focused on two types of inducible transcriptional promoter—Xyls and pBAD. The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design). The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The pBAD promoter exhibits an almost all-or-none behavior upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Based on these research findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD.
As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006/Contains BBa_K3202029) which suggest the two inducible transcription system function properly.
III.Experiment & Results
In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication.
Two inducers with their respective promoters (AraC & pBAD; Xyls & Pm) are coupled with sfGFP to see if there is actually expression leakage when inducer is present quantitatively through flow cytometry. We tested fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration. Theoretically when inducers (AraC and 3MBz ) are present, promoters (pBAD and Pm) are initiated therefore both sfGFP are expressed; while sfGFP shouldn’t be expressed if inducer is absent. When measured at the end of 240 minutes without inducer, BN009/Contains BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/Contains BBa_K3202029(backbone without GFP). BN007/Contains BBa_K3202030(pBAD+sfGFP) also showed a leakage of 90 a.u. The result suggested that both system have noticeable leakage and Xyls has much higher leakage.
When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/Contains BBa_K3202029 stays at a constant level, whereas the fluorescence of both Xyls and pBAD have increased exponentially. This is another sign that both system function properly.
We found an intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/Contains BBa_K3202030 is 200 a.u. higher than BN006/Contains BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone. Same thing happened to BN009/Contains BBa_K3202031. Therefore, in the subsequent experiments, systems are induced to up to 4 hours to ensure the accuracy of the experiment. Results shown above verified the presence of expression leakage of both systems when inducer is not expressed through the detectable fluorescence of reporter protein using flow cytometry.
Bistable System Module
I.Background
We are inspired by a lately published article written by Belén Calles, Angel Goñi-Moreno, and Víctor de Lorenzo to design a double-bistable system which achieves zero basal expression of gene of interest in the absence of any inducer while ensuring constant transcriptional capacities and induction rates among promoters.
“The mechanism of a bistable system is elaborated below: a repressor transcriptionally inhibits the promoter which is responsible for the transcription of sRNA, while the sRNA translationally inhibits the repressor. The gene of interest is translationally coupled to the repressor gene. That is to say, the repressor protein R targets the strong promoter which the inhibitory sRNA is produced, so that R and sRNA are mutually inhibitory. Such arrangement helps to minimize the basal activity of the inducible module and thus suppress leaky gene expression levels.”
II.Design
In our system(Fig. 1), we have transcriptional repressor R1 expressed through the inducible promoter but translationally inhibited by MicC sRNA1 since the RBS of R1 is directly inhibited by sRNA while they initiates the transcription of the two repressors; the gene of interest is translationally coupled to the repressor gene while three promoters which produces the inhibitory sRNA are targeted by their corresponding repressors. Therefore, repressors and sRNAs are mutually inhibitory.
RBS1
The control of the recombinase in coordination with the upstream repressor gene, which is the target of the sRNA, requires a translational coupler cassette. “An efficient mechanism to reach this goal is based on the ability of translating ribosomes to unfold mRNA secondary structures. In our design, translational coupling is achieved by occluding the RBS of gene of interest by formation of a secondary mRNA structure, containing a His-tag sequence added to the 3 ́end of the repressor gene. The hairpin structure prevents the ribosomal recruitment to the recombinase and therefore its translation. In contrast, when the upstream repressor mRNA is actively translated, the 70s ribosome disrupts inhibitory mRNA secondary structure in the downstream gene translation initiation region thus allowing its expression.”
However, during the experiment we tested that the RBS1 (Fig. 2) sequence provided by the original article fails to function in our circuit, we found RBS2 (Fig. 3) with a different working mechanism. The structure of AUGA in RBS2 acts as both the stop codon (AUG) of the former gene sequence, releasing the former peptide chain; takes a step (gene code) backward, pinpointing a new ribosome binding site, and initiated the translation of next gene sequence with the start codon (UGA). However, finally we still used RBS1 since the crux actually lied in another place.
sRNA
Small non‐coding RNAs (sRNAs) have important functions as genetic regulators in prokaryotes. “sRNAs act post‐transcriptionally through complementary pairing with target mRNAs to regulate protein expression. Antisense sRNAs negatively regulates proteins by destabilizing the target protein's mRNA. Antisense sRNAs prevent translation by binding to the target mRNAs in a process mediated by the RNA chaperone Hfq.” On binding, both the mRNAs and sRNAs are degraded, suggesting that prokaryotic sRNAs—unlike their eukaryotic counterparts—act stoichiometrically on their targets.
Repressor
“A DNA-binding repressor blocks the attachment of RNA polymerase to the promoter, thus preventing transcription of the genes into messenger RNA.”
We assume that when the whole system is applied to a factory, the situation where inducer is present is in the working condition of the bacterium. Within one single layer of the bistable system, the transcriptional factor is turned on, Pm initiates the expression of R1, which raises the concentration of R1 and thereby exerts greater inhibition on PR1 and therefore decreases the concentration of sRNA1. As sRNA1 is inhibited the inhibitory effect it exerts on R1 accordingly decreases, therefore the recombinase is expressed through the first layer, and vice versa. Nevertheless, given that the expression of R1 inhibits PR1, even if the recombinase is expressed PR1 can be flipped over but cannot express the second layer. Since there is no further input signal to the second layer of the bistable system, the inhibitory effect of sRNA2 dominates over R2, therefore, both R2 and the toxin cannot be expressed. That is to say, the additional second layer ensures that there is no leakage of toxin when inducer is present.
However, when the bacterium is stolen and thereby inducer isn’t present, the transcriptional factor is turned off, promoter Pm doesn’t work. Therefore, sRNA1 dominates over R1 and then the recombinase is not expressed in the first layer. Since there is a decrease in the expression of R1, the inhibitory effect it exerts on PR1 decreases, therefore PR1 which has already been flipped over when inducer is present (working condition) initiates the expression of R2. Applying the same logic, finally R2 dominates over sRNA2 and expresses the toxin to kill the cell. (Fig. 4)
Why double layer?
The reason why we designed such double-bistable system can be attributed to the consideration that the double-decked bistable systems of both recombinase and toxin could further reinforce the insurance of zero expression of toxin.
When the second bistable system coupling the toxin with the first layer of recombinase, double insurance of zero expression of recombinase and toxin is achieved. However, as the diagram shown below, a single bistable system where toxin is coupled without the second layer of insurance could collapse if the mutual inhibitory effect of repressor and sRNA within the first layer malfunctions. Whereas such concern could be eliminated by the second layer of toxin with a bistable system around.
III. Experiment & Results
After the circuit (Fig. 5) including only one layer of the bistable system is constructed with recombinase being replaced by sfGFP, LacI & TetR serving as repressor, and used 3MBz & AraC as inducers, we qualitatively tested the fluorescence of the reporter protein under two situations when excited blue light. Theoretically, sfGFP should be expressed when inducer is present since LacI, which transcribes sfGFP, dominates over the inhibitory force of sRNA. However, in the experiments when inducers are present and absent, there is no fluorescence of sfGFP (BN027/Contains BBa_K3202074 & 28/Contains BBa_K3202075 & 29/Contains BBa_K3202076 & 30/Contains BBa_K3202077). We deduced that there must be defects lying in our circuit.
Analyzing through the whole circuit, we deduced that the crux might lie in RBS1 for the sake of its overwhelmingly strong hair-pin structure that even if the 3’ end of LacI is translated the ribosomes produced still couldn’t melt sRNA secondary structure, thereby sfGFP cannot be translated. Therefore, we replaced RBS1 with a commonly used RBS: B0034 with weaker strength in order to ensure that there is no problem lying in other sites in the circuit. We tested the sfGFP’s fluorescence in inducers’ presence under excitation of blue light. The expression of sfGFP in the second attempt (Fig. 10B, Fig. 11B & D) proved to us that there’s nothing wrong with the rest of the circuit except for RBS1.
Through further searching we found that RBS2 has a weaker hair pin structure while it could support the translation of repressor. We replaced RBS B0034 with RBS2. Results are shown in the two graphs below. Plasmid with RBS2 expressed sfGFP with quite a high expression leakage when both inducers are absent. We thought that such high expression leakage might be the result of the weak hair-pin structure of RBS2, which means that we still need a strong RBS to reduce expression leakage. Hence, by we were inspired to realize that we might wrongly include the terminator of LacI before RBS. If we include the terminator, the transcription of LacI stops there and therefore the LacI protein produced by translation ends in front of RBS and leaves the mRNA secondary structure before melting it, sfGFP failed to be expressed.
As shown in Fig. 15 & 16 from flow cytometry experiments: at the same time period, the conc. of inducer increases from 0.000 to 1.000 mMol, BN006/Contains BBa_K3202029, which carries no sfGFP, has a low fluorescence level around 0.60 a.u. in inducer’s absence. BN007/Contains BBa_K3202030 and BN009/Contains BBa_K3202031 without the control of bistable system had their fluorescence value at a relatively higher level in inducer’s absence and their fluorescence value increase as conc. of inducers increases. BN043/Contains BBa_K3202048 & 51/Contains BBa_K3202051 & 45/Contains BBa_K3202049 & 49/Contains BBa_K3202050 (under the control of bistable system) have their fluorescence value lower than that of BN007/Contains BBa_K3202030 and BN009/Contains BBa_K3202031 in inducer’s absence and have their fluorescence value as conc. of inducer increases.
As shown in Fig. 17 & 18 from flow cytometry experiments: in the absence of inducer at the indicated time points (4h and 24h after induction), BN006/Contains BBa_K3202029, which carries no sfGFP, has a low fluorescence level around 0.60 a.u. in inducer’s absence. BN043/Contains BBa_K3202048 & 51/Contains BBa_K3202051 & 45/Contains BBa_K3202049 & 49/Contains BBa_K3202050 (under the control of bistable system) have their fluorescence value higher than that of BN006/Contains BBa_K3202029 while lower than that of BN007/Contains BBa_K3202030 &BN009/Contains BBa_K3202031 (without the control of bistable system) in inducer’s absence.
We improved our gene circuit upon that reflection, with exclusion of the terminator of repressors and replacement of BRS2 using RBS1, and made the third trial. The results are shown below.
As shown in Fig. 20 & 21 from flow cytometry experiments: in the absence of inducer at the indicated time points (4h and 24h after induction), BN006/Contains BBa_K3202029, which carries no sfGFP, has a low fluorescence level around 0.60 a.u. in inducer’s absence. BN007/Contains BBa_K3202030 and BN009/Contains BBa_K3202031are two plasmids without the control of bistable system; they displayed high fluorescence level as time passed. BN054/Contains BBa_K3202044 and BN055/Contains BBa_K3202045 have similarly no fluorescence level as BN006/Contains BBa_K3202029.
As shown in Fig. 22 & 23 from flow cytometry experiments: at same time period, as the conc. of inducers increases from 0.0001 (nearly no inducer) to 10 mMol, BN006/Contains BBa_K3202029, which carries no sfGFP, always has a low fluorescence level around 0.60 a.u. While both two plasmids (BN054/Contains BBa_K3202044, induced by Ara; BN055/Contains BBa_K3202045, induced by 3MBz) have their fluorescence level increase from around 10 a.u. (lack of expression of sfGFP) when inducer is absent to a higher value.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 93
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 815
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 278