Difference between revisions of "Part:BBa K1141000"
PeterGockel (Talk | contribs) |
PeterGockel (Talk | contribs) |
||
| (7 intermediate revisions by the same user not shown) | |||
| Line 9: | Line 9: | ||
<p align="center" id="legend">Figure 1: Excitation (left peak) and Emission (right peak) spectra for mCherry.</p> | <p align="center" id="legend">Figure 1: Excitation (left peak) and Emission (right peak) spectra for mCherry.</p> | ||
| − | + | <h1> Translation Enhancing 5'-UTR (Improvement by iGEM TU_Darmstadt 2019) </h1> | |
| + | <html> | ||
| + | <script id="MathJax-script" async | ||
| + | src="https://2019.igem.org/wiki/index.php?title=Template:TU_Darmstadt/MathjaxJS&action=raw&ctype=text/javascript"></script> | ||
| + | |||
| + | <div class="container"> | ||
| + | <div class="row"> | ||
| + | <div class="col mx-2"> | ||
| + | <h3>Usage and Biology</h3> | ||
| + | <hr class="head"> | ||
| + | <p> | ||
| + | |||
| + | Improved version: <a | ||
| + | href="https://parts.igem.org/Part:BBa_K3187014" | ||
| + | target="_blank"> BBa_K3187014</a> | ||
| + | |||
| + | <br></br> | ||
| + | The part was improved in terms of its expression level, based on the insertion of a 5’-untranslated region (5’-UTR) upstream of the coding sequence. This 5'-UTR was adapted from iGEM Bielefeld 2015 ( <a | ||
| + | href="https://parts.igem.org/Part:BBa_K1758100" | ||
| + | target="_blank"> BBa_K1758100</a>) and is based on the research of Olins <i>et</i> al | ||
| + | <sup id="cite_ref-1" class="reference"> | ||
| + | <a href="#cite_note-1">[1] </a> | ||
| + | </sup> | ||
| + | and Takahashi <i>et</i> al. | ||
| + | <sup id="cite_ref-2" class="reference"> | ||
| + | <a href="#cite_note-2">[2] </a> | ||
| + | </sup>. | ||
| + | It contains the strong ribosomal binding site (RBS) g10-L from the T7 bacteriophage and a sequence that plays a role in the regulation of mRNA binding to and release from the 30S ribosomal subunit. The 5'-UTR therefore enhances the translation efficiency of the following coding sequence (CDS) (Fig. 1). | ||
| + | |||
| + | </p> | ||
| + | |||
| + | <img | ||
| + | class="img-fluid center" | ||
| + | src="https://2019.igem.org/wiki/images/6/62/T--TU_Darmstadt--RibosomLiberation.png" | ||
| + | style="max-width:80%" | ||
| + | /> | ||
| + | <div class="caption"> | ||
| + | <p> | ||
| + | <b> | ||
| + | <center> Figure 1: | ||
| + | </b> Schematic depiction of the composition and interaction of the enhancer sequence with the 30S ribosomal subunit described by Takahashi <i>et.</i> al. <sup id="cite_ref-2" class="reference"> | ||
| + | <a href="#cite_note-2">[2] </a> | ||
| + | </sup>. | ||
| + | </center> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <p> | ||
| + | The sequence of the translation enhancing 5’-UTR can be divided into the four main features listed below: | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | <img | ||
| + | class="img-fluid center" | ||
| + | src="https://2019.igem.org/wiki/images/f/f3/T--TU_Darmstadt--EnhancerInColor.png" | ||
| + | style="text= align; max-width:85%" | ||
| + | /> | ||
| + | |||
| + | |||
| + | <br></br> | ||
| + | <div class="container-noborders"> | ||
| + | <div class="table-responsive-sm"> | ||
| + | <table class="table table-light"> | ||
| + | <tr> | ||
| + | <th scope="col">Sequence</th> | ||
| + | <th scope="col">Function</th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="color:#d59b9b; word-wrap:break-word;"> | ||
| + | AATAATTTTGTT<br />TTAACTTTAA | ||
| + | </td> | ||
| + | <td> | ||
| + | The T7 g10 leader sequence (first described by Olins <i>et</i> al<sup id="cite_ref-1" class="reference"> | ||
| + | <a href="#cite_note-1">[1] </a> | ||
| + | </sup>)increases the efficiency of translation initiation. This sequence contains the epsilon motif TTAACTTTA which enhances the binding of the mRNA to the 16S rRNA. | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="color:#a5c8c8;">poly-A</td> | ||
| + | <td> | ||
| + | Referring to Takahashi et al.<sup id="cite_ref-2" class="reference"> | ||
| + | <a href="#cite_note-2">[2] </a> | ||
| + | </sup> a spacer between the epsilon motive and the RBS improves the translation rate. | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="color:#698888;">GAAGGAG</td> | ||
| + | <td> | ||
| + | According to Karig <i>et</i> al.<sup id="cite_ref-3" class="reference"> | ||
| + | <a href="#cite_note-3">[3] </a> | ||
| + | </sup> and Lentini et. al<sup id="cite_ref-4" class="reference"> | ||
| + | <a href="#cite_note-4">[4] </a> </sup> a distance of 4-9 bases between RBS and start codon increases the translation efficiency. | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="color:#cc9966;">AATAATCT</td> | ||
| + | <td> | ||
| + | According to Lentini et. al<sup id="cite_ref-4" class="reference"> | ||
| + | <a href="#cite_note-4">[4] </a> </sup> an AT-rich composition between the RBS and the start codon results in the best expression results. | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </div> | ||
| + | </div> | ||
| + | <h3>Results</h3> | ||
| + | <hr class="head"> | ||
| + | <p> | ||
| + | Both the original and the improved version were cloned in pSB1C3. <i>E. coli</i> BL21 (DE3) was transformed using BBa_K1073024 and cultured using 25 mL LB medium supplemented with Chloramphenicol (based on the pSB1C3 resistance). The expression was measured (Ex: 587 nm, Em: 610 nm) over a period of 6 hours after induction with 0.5 mM IPTG (<b>Fig. 2</b>) using a SpectraMax M5E. | ||
| + | </p> | ||
| + | <img | ||
| + | class="img-fluid center" | ||
| + | src="https://2019.igem.org/wiki/images/e/e7/T--TU_Darmstadt--Improved_BBa_K1141001.png" | ||
| + | style="max-width:80%" | ||
| + | /> | ||
| + | <div class="caption"> | ||
| + | <p> | ||
| + | <b> | ||
| + | <center> Figure 2: | ||
| + | </b>Expression of mCherry with (improved BBa_K1141000) and without (BBa_K1141000) the translation enhancing 5’ untranslated region was measured (Ex: 587 nm, Em: 610 nm) over a period of 6 hours after induction with 0.5 mM IPTG using a SpectraMax M5E.</center> | ||
| + | </p> | ||
| + | </div> | ||
| + | <p> | ||
| + | <b>Figure 2</b> shows an increased expression of the improved BBa_K1141001 in comparison to BBa_K1141001. The increased expression rate of the improved version (left) after 6 hours can also be observed with the naked eye (<b>Fig. 3</b>). | ||
| + | </p> | ||
| + | |||
| + | <img | ||
| + | class="img-fluid center" | ||
| + | src="https://2019.igem.org/wiki/images/7/72/T--TU_Darmstadt--mCherryImprovement.png" | ||
| + | style="max-width:50%" | ||
| + | /> | ||
| + | |||
| + | <div class="caption"> | ||
| + | <p> | ||
| + | <b> | ||
| + | <center> Figure 3: | ||
| + | </b><I>E. coli</i> BL21 (DE3) cultures transformed with BBa_K1141000 (right) and the improved version of BBa_ K1141000 (left) after cultivation in LB medium supplemented with Chloramphenicol (according to the resistance provided by pSB1C3) for 6 hours. </center> | ||
| + | </p> | ||
| + | </div> | ||
| + | |||
| + | <h2>References</h2> | ||
| + | <ol class="references"> | ||
| + | <li id="cite_note-1"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-1">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | Peter O. Olins and Shaukat H. Rangwala, A Novel Sequence Element Derived from Bacteriophage T7 mRNA Acts as an Enhancer of Translation of the lacZ Gene in Escherichia coli, The Journal of Biological Chemistry, Vol. 264, No. 29, Issue of Ovtobet 15, pp. 16973-16976, 1989 | ||
| + | <a rel="nofollow" class="external autonumber" href=" https://www.ncbi.nlm.nih.gov/pubmed/2676996" | ||
| + | target="_blank">[1] </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | <li id="cite_note-2"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-2">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | Shuntaor Takahashi, Hiroyuki Furusawa, Takuya Ueda and Yoshio Okahata, Translation Enhancer Improves the Ribosome Liberation from Translation Initiation, J. Am. Chem. Soc. 2013, 135 35, 13096-13106 | ||
| + | <a rel="nofollow" class="external autonumber" | ||
| + | href=" https://www.ncbi.nlm.nih.gov/pubmed/23927491 | ||
| + | " target="_blank">[2] | ||
| + | </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | <li id="cite_note-3"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-3">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | David K. Karig, Sukanya Iyer, Michael L. Simpson and Mitchel J. Doktycz, Expression optimization and synthetic gene networks in cell-free systems, Nucleic Acids Res. 2012 Apr; 40(8): 3763.3774 | ||
| + | <a rel="nofollow" class="external autonumber" | ||
| + | href="https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3333853/" target="_blank">[3] | ||
| + | </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | <li id="cite_note-4"> | ||
| + | <span class="mw-cite-backlink"> | ||
| + | <a href="#cite_ref-4">↑</a> | ||
| + | </span> | ||
| + | <span class="reference-text"> | ||
| + | Roberta Lentini, Silvia Perez Santero, Fabio Chizzolini, Dario Cecchi, Jason Fontana, Marta Marchioretto, Christina Del Bianco, Jessica L. Terrell, Amy C. Spencer, Laura Martini, Michele Forlin, Michael Assfalg, Mauro Dalla Serra, William E. Bentley and Sheref S. Mansy, Integrating artificial with natural cells to translate chemical messages that direct E. coli behavior, Nature Communications 5, Article number: 4012 (2014) | ||
| + | <a rel="nofollow" class="external autonumber" | ||
| + | href=" https://www.nature.com/articles/ncomms5012 " | ||
| + | target="_blank">[4] </a> | ||
| + | </span> | ||
| + | </li> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | </ol> | ||
| + | |||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | </div> | ||
| + | <br></br> | ||
</html> | </html> | ||
| + | |||
| + | |||
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
Latest revision as of 14:59, 20 October 2019
Plac-RBS-mCherry-double terminator (IPTG-inducible)
This part contains all the necessary DNA to produce the red fluorescent protein mCherry with the PLac promoter. PLac repression is low in most E. coli strains and the promoter will be leaky. Efficient repression can be implemented by additionally transforming a strain with the pREP4 plasmid from the M15 E. coli strain of QIAGEN, or transforming the part into this strain. The figure below shows the excitation and emission spectra as can be obtained from CHROMA's spectra viewer:

Figure 1: Excitation (left peak) and Emission (right peak) spectra for mCherry.
Translation Enhancing 5'-UTR (Improvement by iGEM TU_Darmstadt 2019)
Usage and Biology
Improved version: BBa_K3187014
The part was improved in terms of its expression level, based on the insertion of a 5’-untranslated region (5’-UTR) upstream of the coding sequence. This 5'-UTR was adapted from iGEM Bielefeld 2015 ( BBa_K1758100) and is based on the research of Olins et al
[1]
and Takahashi et al.
[2]
.
It contains the strong ribosomal binding site (RBS) g10-L from the T7 bacteriophage and a sequence that plays a role in the regulation of mRNA binding to and release from the 30S ribosomal subunit. The 5'-UTR therefore enhances the translation efficiency of the following coding sequence (CDS) (Fig. 1).
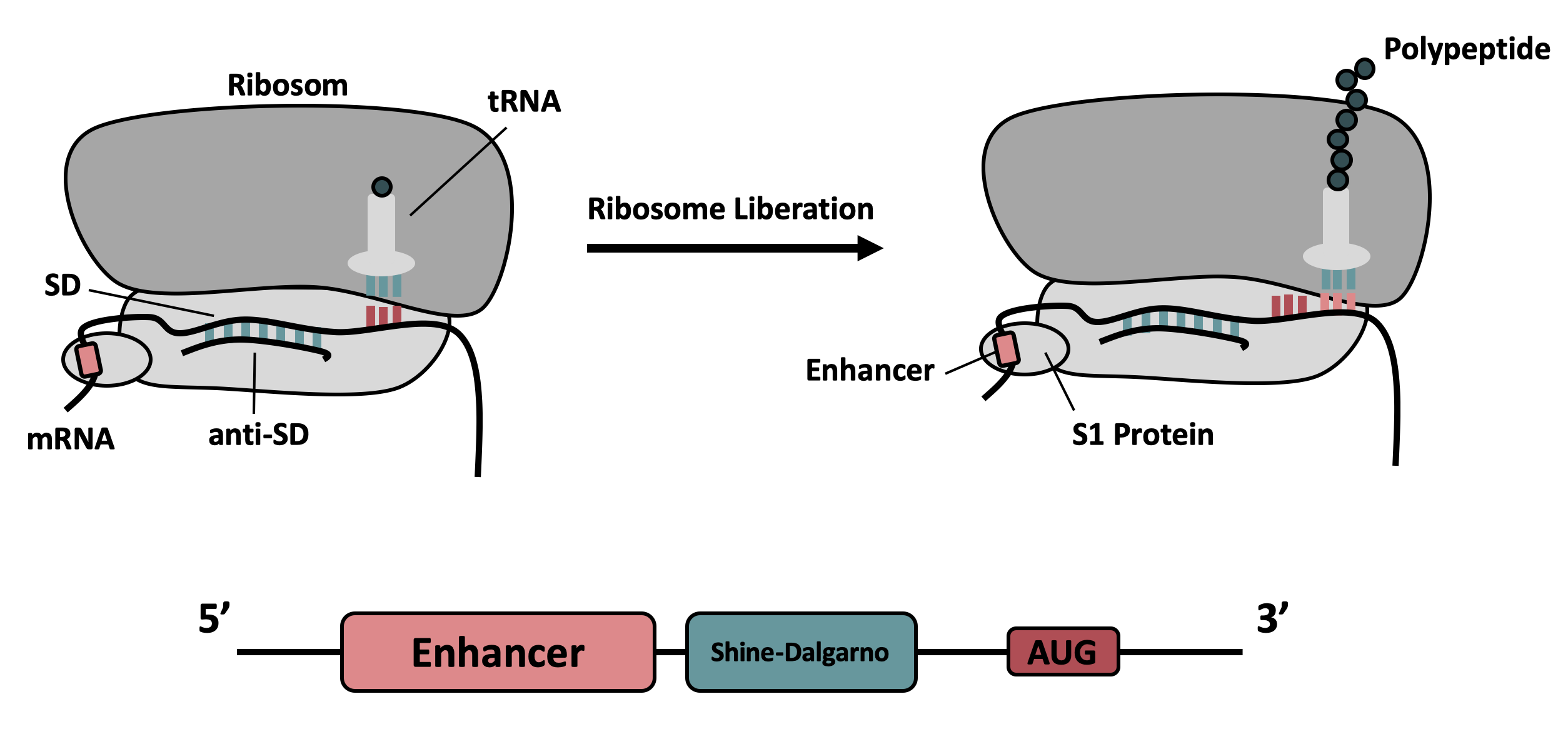
The sequence of the translation enhancing 5’-UTR can be divided into the four main features listed below:

| Sequence | Function |
|---|---|
|
AATAATTTTGTT TTAACTTTAA |
The T7 g10 leader sequence (first described by Olins et al [1] )increases the efficiency of translation initiation. This sequence contains the epsilon motif TTAACTTTA which enhances the binding of the mRNA to the 16S rRNA. |
| poly-A | Referring to Takahashi et al. [2] a spacer between the epsilon motive and the RBS improves the translation rate. |
| GAAGGAG | According to Karig et al. [3] and Lentini et. al [4] a distance of 4-9 bases between RBS and start codon increases the translation efficiency. |
| AATAATCT | According to Lentini et. al [4] an AT-rich composition between the RBS and the start codon results in the best expression results. |
Results
Both the original and the improved version were cloned in pSB1C3. E. coli BL21 (DE3) was transformed using BBa_K1073024 and cultured using 25 mL LB medium supplemented with Chloramphenicol (based on the pSB1C3 resistance). The expression was measured (Ex: 587 nm, Em: 610 nm) over a period of 6 hours after induction with 0.5 mM IPTG (Fig. 2) using a SpectraMax M5E.

Figure 2 shows an increased expression of the improved BBa_K1141001 in comparison to BBa_K1141001. The increased expression rate of the improved version (left) after 6 hours can also be observed with the naked eye (Fig. 3).

References
- ↑ Peter O. Olins and Shaukat H. Rangwala, A Novel Sequence Element Derived from Bacteriophage T7 mRNA Acts as an Enhancer of Translation of the lacZ Gene in Escherichia coli, The Journal of Biological Chemistry, Vol. 264, No. 29, Issue of Ovtobet 15, pp. 16973-16976, 1989 [1]
- ↑ Shuntaor Takahashi, Hiroyuki Furusawa, Takuya Ueda and Yoshio Okahata, Translation Enhancer Improves the Ribosome Liberation from Translation Initiation, J. Am. Chem. Soc. 2013, 135 35, 13096-13106 [2]
- ↑ David K. Karig, Sukanya Iyer, Michael L. Simpson and Mitchel J. Doktycz, Expression optimization and synthetic gene networks in cell-free systems, Nucleic Acids Res. 2012 Apr; 40(8): 3763.3774 [3]
- ↑ Roberta Lentini, Silvia Perez Santero, Fabio Chizzolini, Dario Cecchi, Jason Fontana, Marta Marchioretto, Christina Del Bianco, Jessica L. Terrell, Amy C. Spencer, Laura Martini, Michele Forlin, Michael Assfalg, Mauro Dalla Serra, William E. Bentley and Sheref S. Mansy, Integrating artificial with natural cells to translate chemical messages that direct E. coli behavior, Nature Communications 5, Article number: 4012 (2014) [4]
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
