Difference between revisions of "Part:BBa K3202029"
Katherine Z4 (Talk | contribs) |
Katherine Z4 (Talk | contribs) |
||
| Line 21: | Line 21: | ||
As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006/BBa_K3202029) which suggest the two inducible transcription system function properly. | As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006/BBa_K3202029) which suggest the two inducible transcription system function properly. | ||
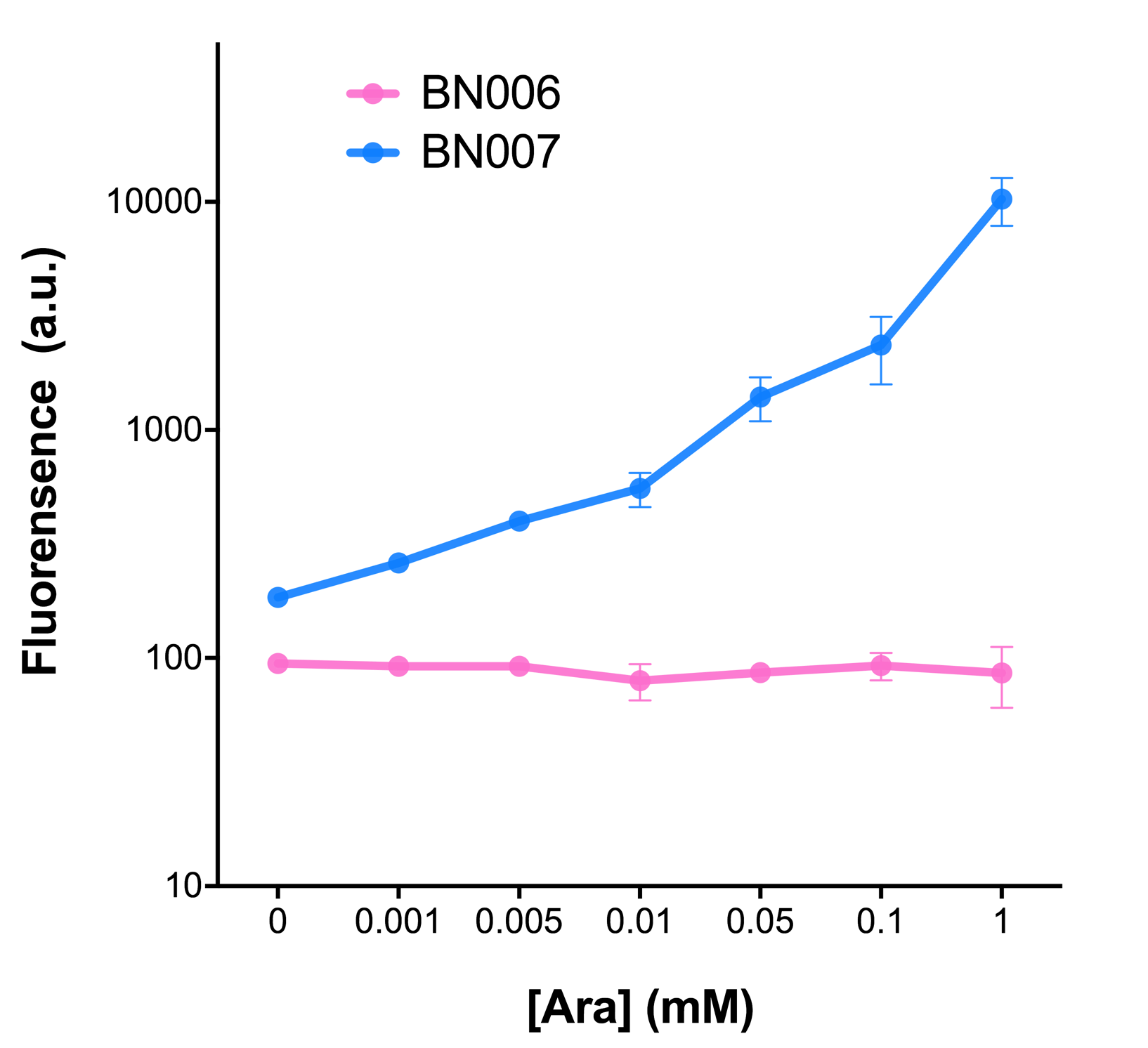
| − | [[File:T--BHSF_ND--Leakage experiment 1.png| | + | [[File:T--BHSF_ND--Leakage experiment 1.png|432px|thumb|left|alt=comparison between BN006/BBa_K3202029 and BN007/BBa_K3202030|Figure 1: pBAD(BN007/BBa_K3202030) induction curve at different concentrations after 4 hours compared to backbone.(BN006/BBa_K3202029)]] |
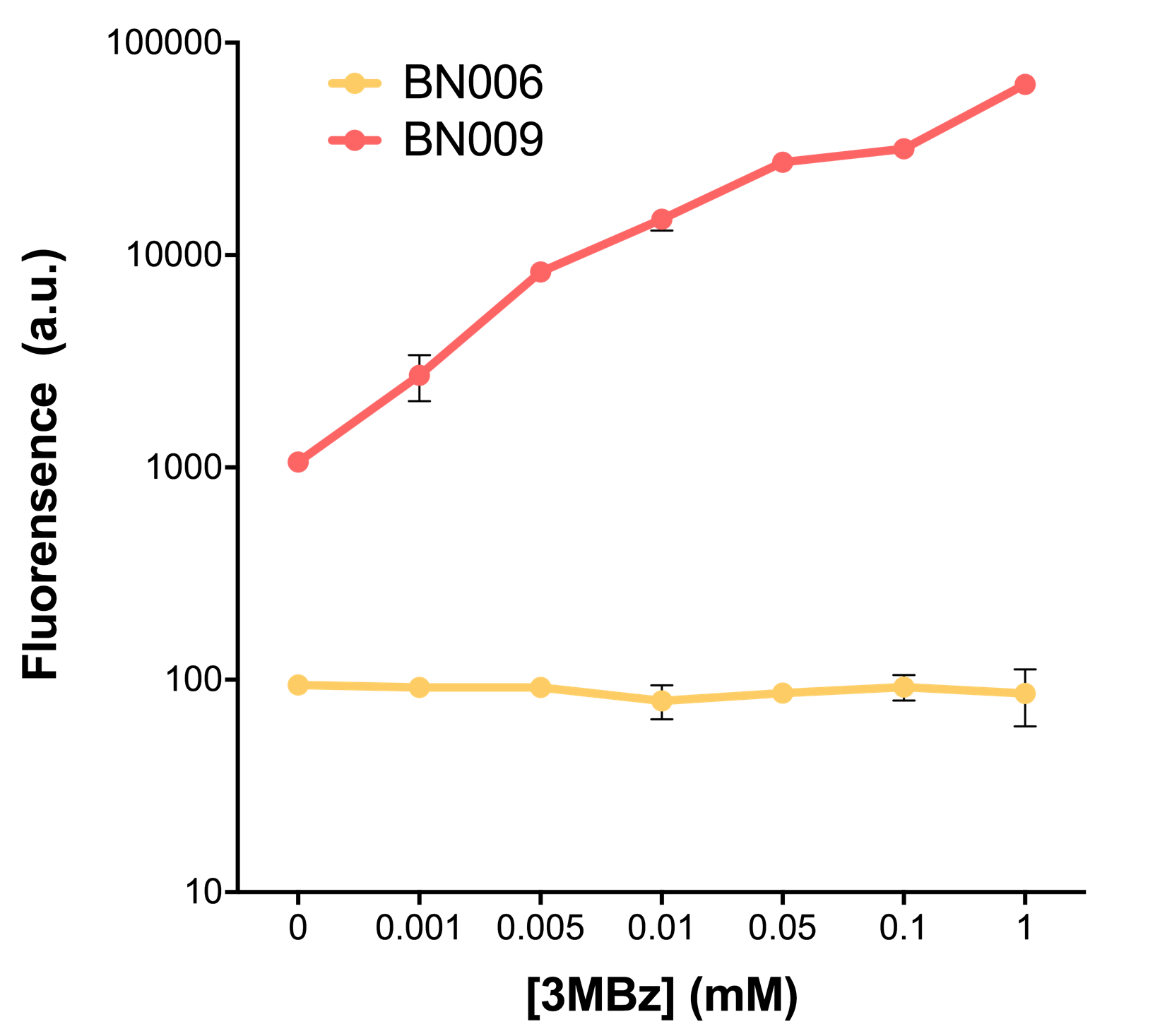
| − | [[File:T--BHSF_ND--Leakage experiment 2.png| | + | [[File:T--BHSF_ND--Leakage experiment 2.png|432px|thumb|right|alt=comparison between BN006/BBa_K3202029 and BN009/BBa_K3202031|Figure 2: Xyls(BN009/BBa_K3202031) induction curve at different concentrations after 4 hours compared to backbone(BN006/BBa_K3202029)]] |
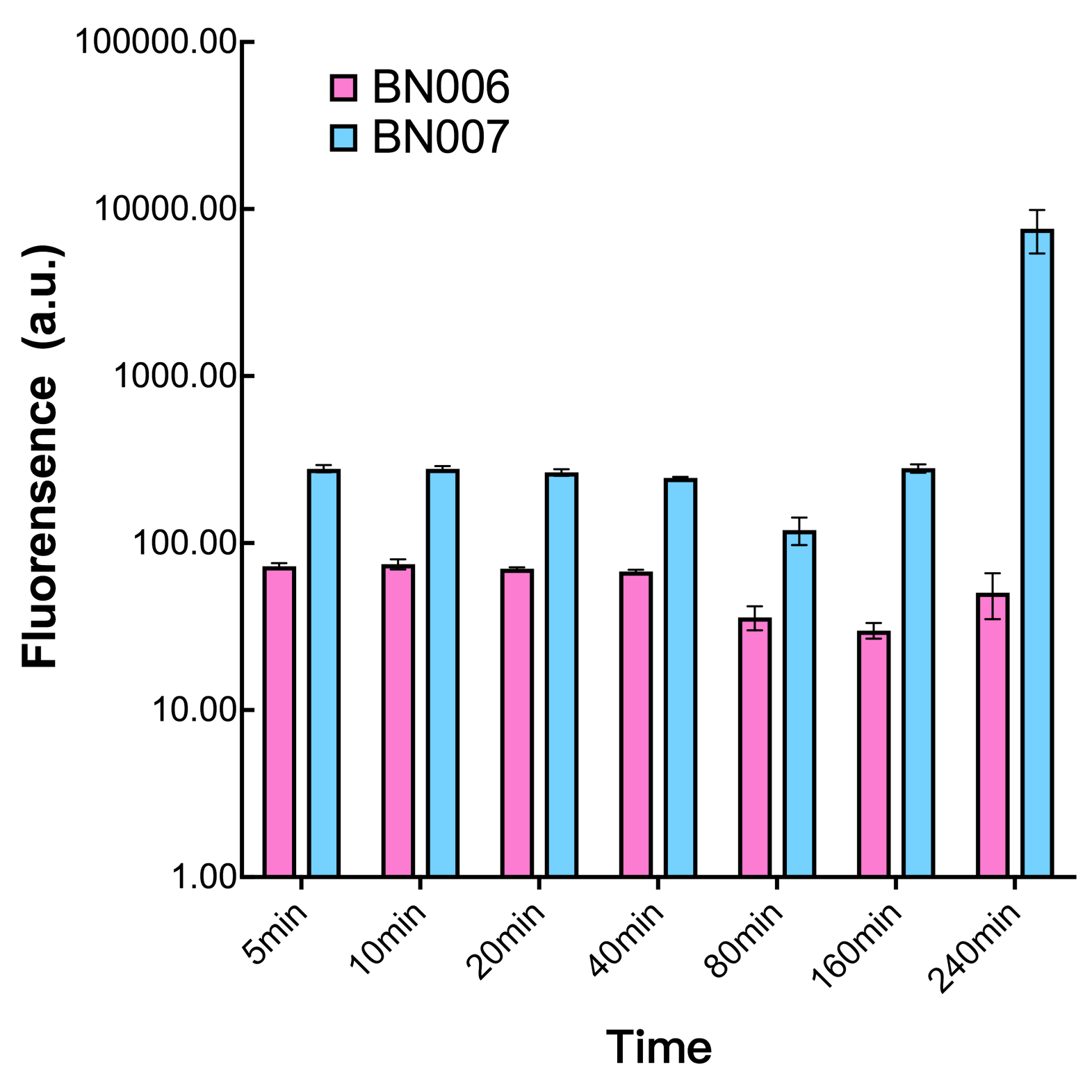
| − | [[File:T--BHSF_ND--Leakage experiment 3.png| | + | [[File:T--BHSF_ND--Leakage experiment 3.png|432px|thumb|left|alt=comparison between BN006/BBa_K3202029 and BN007/BBa_K3202030|Figure3: Time induction curve pBAD(BN007/BBa_K3202030) at the concentration of 1mM compared to backbone (BN006/BBa_K3202029)]] |
| + | |||
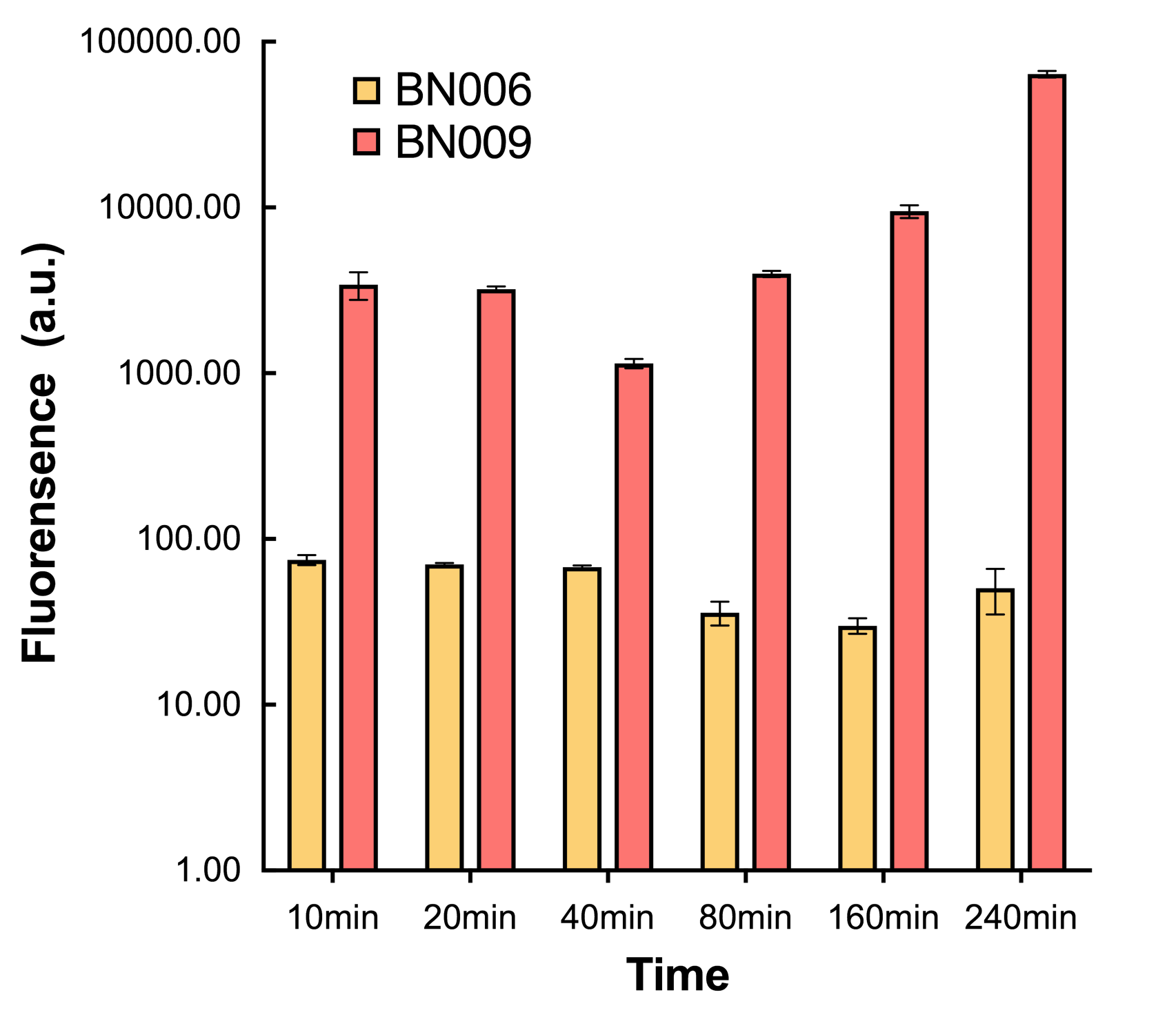
| + | [[File:T--BHSF_ND--Leakage experiment 4.png|432px|thumb|right|alt=comparison between BN006/BBa_K3202029 and BN009/BBa_K3202031|Figure4: Time induction curve for Xyls(BN009/BBa_K3202031) at the concentration of 1mM compared to backbone (BN006/BBa_K3202029)]] | ||
| − | |||
=== Experiment & Results === | === Experiment & Results === | ||
Revision as of 14:44, 20 October 2019
Pc-Xyls
This composite part is designed as a genetic circuit that is used as a control group of our later experiment.
This acts as a control group in our experiment. It is compared to BBa_K3202030 and BBa_K3202031.
Leakage/Inducer Module
Background
Gene expression depends largely on transcription and translation. Such processes display activated/non-activated states. For some situations, we would expect zero expression of GOI without the presence of inducer. However, leakage still exist which is due to the incomplete OFF stage of the transcriptional system. Even without proper inducers, GOI is still partly expressed in some case. Such leakage is called the basal leakage.In some cases, as long as one molecule of toxin protein, is translated, it can kill the entire cell. Thus, preventing leakage is a vital in regulating gene expression. Current methods of solving leakage problems includes introducing strong inducible promoters to the system. However, there are substantially no regulating system or transcriptional factor that can function as a technically ON/OFF switch to prevent basal level leakage. This may due to weak RNAP-DNA interactions or different transcriptional capacities and induction rate of different promoters and inducers.
Design
We mainly focused on two types of inducible transcriptional promoter—Xyls and pBAD. The Xyls protein is the positive transcription regulator of the TOL plasmid meta-cleavage pathway operon Pm. Xyls belongs to the AraC family of transcriptional regulators and exhibits an N-terminal domain involved in effector recognition, and a C-terminal domain.The activator Xyls is usually is usually activated by the benzoate and its derivatives (3MBz in our design). The pBAD promoter holds great potential to control the expression of genes that require tight regulation, as it is capable of both an induction and repression. The pBAD promoter exhibits an almost all-or-none behavior upon induction with arabinose when located on a high copy vector, but allows for gradual induction when cloned into a low copy vector. Based on these research findings, a low copy vector was used to investigate the ability to inhibit gene expression subsequent to induction of pBAD.
As the result shows, both system exhibit relative high level of fluorescence compared to our backbone design( BN006/BBa_K3202029) which suggest the two inducible transcription system function properly.
Experiment & Results
In our experiment of leakage testing, we used low-copying plasmid backbone PSB4K5, with a PSC101 origin of replication.
Two inducers with their respective promoters (Arac & pBAD; Xyls & pm) are coupled with sfGFP to see if there is actually expression leakage when inducer is present quantitatively through flow cytometry. We tested fluorescence under different inducer concentration at the given time of 240 minutes and under different time stage at a given inducer concentration. Theoretically when inducers (AraC and 3MBz ) are present, promoters (pBAD and Pm) are initiated therefore both sfGFP are expressed; while sfGFP shouldn’t be expressed if inducer is absent. When measured at the end of 240 minutes without inducer, BN009/BBa_K3202031(Xyls+sfGFP) has a basal leakage of 968 a.u. compared to BN006/BBa_K3202029(backbone without GFP). BN007/BBa_K3202030(pBAD+sfGFP) also showed a leakage of 90 a.u. The result suggested that both system have noticeable leakage and Xyls has much higher leakage.
When we gradually increased inducer concentration to 1mM, the fluorescence of BN006/BBa_K3202029 stays at a constant level, whereas the fluorescence of both Xyls and pBAD have increased exponentially. This is another sign that both system function properly.
We found a intriguing result when measuring the fluorescence of the two system under a constant inducer concentration of 1mM. During the first 160 minutes, the fluorescence of BN007/BBa_K3202030 is 200 a.u. higher than BN006/BBa_K3202029. However, at 240 minutes, the fluorescence value suddenly increased to 7641 a.u., which is 7590 a.u., higher than the backbone. Same thing happened to BN009/BBa_K3202031. Therefore, in the subsequent experiments, systems are induced to up to 4 hours to ensure the accuracy of the experiment. Results shown above verified the presence of expression leakage of both systems when inducer is not expressed through the detectable fluorescence of reporter protein using flow cytometry.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 93
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal NgoMIV site found at 815
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 278