Difference between revisions of "Part:BBa K2997008"
| (29 intermediate revisions by 3 users not shown) | |||
| Line 5: | Line 5: | ||
===Background=== | ===Background=== | ||
| − | As part of our project, we decided to make a device that can sense <i>p</i>-Cresol. According to our research, we have discovered a group of bacteria known as <i>Pseudomonas</i> | + | As part of our project, we decided to make a device that can sense <i>p</i>-Cresol. According to our research<sup>[1]</sup>, we have discovered a group of bacteria known as <i>Pseudomonas</i> are able to sense <i>p</i>-Cresol naturally. In a previous study<sup>[2]</sup>, the ability of <i>Pseudomonas</i> to sense <i>p</i>-Cresol is used by them to detect <i>p</i>-Cresol in the environment. |
| − | + | ||
===Part Construction & Experimental Results=== | ===Part Construction & Experimental Results=== | ||
| + | <html> | ||
| + | The <i>pchR</i>-GFP was constructed from IDT DNA synthesis in the form of 3 gBlock gene fragments which contains <a href="https://parts.igem.org/Part:BBa_K2997003">BBa_K2997003</a> and <a href="https://parts.igem.org/Part:BBa_K2997004">BBa_K2997004</a> via overhang PCR. Then, we performed digestion on the finished part and ligated it into the pSB1C3 vector. Subsequently, we transformed the <i>pchR</i>-GFP construct into <i>E. coli</i> Top 10 using electroporation. After allowing the transformed <i>E. coli</i> Top 10 to grow on LB with Cm (Chloramphenicol) plates, colony PCR and sequencing was carried out to determine successful clones. The colony PCR results indicated that the <i>E. coli</i> has pSB1C3 with <i>pchR</i>-GFP. However, after several attempts to construct <i>pchR</i>-GFP, all the sequencing results indicated mutations occurred on all of our samples. | ||
| + | </html> | ||
| − | + | <html> | |
| + | <br> | ||
| + | <div style="width=100%; display:flex; align-items: center; justify-content: center;"> | ||
| + | <img src="https://static.igem.org/mediawiki/parts/b/bd/PchR_sequencing_result3.jpg" style="width:90%;"> | ||
| + | </div> | ||
| + | <br> | ||
| + | </html> | ||
| − | << | + | <html> |
| + | <center> | ||
| + | Fig. 15. Sequencing results of the pSB1C3 w/ <i>pchR</i>-GFP construct. | ||
| + | </center> | ||
| + | <br> | ||
| + | </html> | ||
| − | + | These results led us to hypothesize that the PchR protein is toxic to our <i>E. coli</i>. Moreover, according to a research paper<sup>[4]</sup> we have found, it is suggested in the study that <i>p</i>-Cresol is toxic to Gram-negative bacteria, which includes <i>E. coli</i>. Hence, this plasmid construct has been dropped. | |
| − | + | Therefore, the construction of <i>pchR</i>-GFP was carried out again by re-doing overlap PCR, where we combine all the three IDT gene fragments into the full-length <i>pchR</i>-GFP construct. For this time, the finished full-length <i>pchR</i>-GFP part was then inserted into the vector pBBR1 MCS-4. | |
| − | |||
<html> | <html> | ||
<br> | <br> | ||
| Line 27: | Line 39: | ||
</html> | </html> | ||
| − | Fig. 16. Gel electrophoresis result of pBBR1 MCS-4 with full length pchR-GFP. Lane 1: Marker; Lane 2: Pre-ligation of vector pBBR1 MCS-4 (4950bp) and insert pchR-GFP (3138bp); Lane 3: Post-ligation of pBBR1 MCS-4 with full pchR-GFP (8098 bp) | + | <html> |
| + | <center> | ||
| + | Fig. 16. Gel electrophoresis result of pBBR1 MCS-4 with full length <i>pchR</i>-GFP. Lane 1: Marker; Lane 2: Pre-ligation of vector pBBR1 MCS-4 (4950bp) and insert <i>pchR</i>-GFP (3138bp); Lane 3: Post-ligation of pBBR1 MCS-4 with full <i>pchR</i>-GFP (8098 bp) | ||
After that, we proceeded into transforming the construct into <i>Pseudomonas fluorescens</i> 55 and <i>Pseudomonas aeruginosa</i> PAD1 via electroporation. | After that, we proceeded into transforming the construct into <i>Pseudomonas fluorescens</i> 55 and <i>Pseudomonas aeruginosa</i> PAD1 via electroporation. | ||
| + | </center> | ||
| + | </html> | ||
<html> | <html> | ||
<br> | <br> | ||
| − | However, due to the complications surrounding the selection marker being used (Ampicillin), we were not able to get a definitive result that indicates that we have positive colonies. This is due to the fact that <i>P.fluorescens</i> 55 and <i>P.aeruginosa</i> PAD1 are naturally resistant to ampicillin and the vector we used for our construct | + | However, due to the complications surrounding the selection marker being used (Ampicillin), we were not able to get a definitive result that indicates that we have positive colonies. This is due to the fact that <i>P.fluorescens</i> 55 and <i>P.aeruginosa</i> PAD1 are naturally resistant to ampicillin and the vector we used for our construct contains ampicillin selection marker. Hence, positive colonies cannot be differentiated from negative colonies. Besides that, our colony PCR results on the single colonies were also proven to be negative. |
| − | < | + | </html> |
| − | Therefore, with time being one of the factors, we decided to drop any further construction and functional experiments surrounding the <i>p</i>-Cresol sensing part (pchR-GFP). | + | |
| + | <html> | ||
| + | Therefore, with time being one of the factors, we decided to drop any further construction and functional experiments surrounding the <i>p</i>-Cresol sensing part (<i>pchR</i>-GFP). | ||
<br> | <br> | ||
</html> | </html> | ||
| − | === | + | ===Mechanism and Biology=== |
| − | The construct is primarily centered upon <i>Pseudomonas fluorescens</i> (PC24), as it has a gene cluster that consists of a positive inducible operon, pchR, which allows it to sense <i>p</i>-Cresol and promote the transcription of the downstream region to degrade the p-Cresol. The pchR gene can be translated into activator proteins that can bind with the inducer (<i>p</i>-Cresol) and leads to the conformation of the protein. As a result, the activator protein binds with <i>p</i>-Cresol and the eventual protein-<i>p</i>-Cresol complex will then bind onto the <i>p</i>-Cresol sensing region that is placed just downstream of pchR and start transcription of the gene downstream. | + | The construct is primarily centered upon <i>Pseudomonas fluorescens</i> (PC24)<sup>[2][3]</sup>, as it has a gene cluster that consists of a positive inducible operon, <i>pchR</i>, which allows it to sense <i>p</i>-Cresol and promote the transcription of the downstream region to degrade the <i>p</i>-Cresol. The <i>pchR</i> gene can be translated into activator proteins that can bind with the inducer (<i>p</i>-Cresol) and leads to the conformation of the protein. As a result, the activator protein binds with <i>p</i>-Cresol and the eventual protein-<i>p</i>-Cresol complex will then bind onto the <i>p</i>-Cresol sensing region that is placed just downstream of <i>pchR</i> and start transcription of the gene downstream. |
<!-- --> | <!-- --> | ||
| Line 56: | Line 74: | ||
<ol type="1"> | <ol type="1"> | ||
<li>Cho, Ah Ra, Eun Jin Lim, Yaligara Veeranagouda, and Kyoung Lee* | <li>Cho, Ah Ra, Eun Jin Lim, Yaligara Veeranagouda, and Kyoung Lee* | ||
| − | Department of Microbiology, Changwon National University, Changwon-si, Kyongnam 641-773, Korea . Identification of a p-Cresol Degradation Pathway by a GFP-Based Transposon in <i>Pseudomonas</i> and Its Dominant Expression in Colonies | + | Department of Microbiology, Changwon National University, Changwon-si, Kyongnam 641-773, Korea . Identification of a <i>p</i>-Cresol Degradation Pathway by a GFP-Based Transposon in <i>Pseudomonas</i> and Its Dominant Expression in Colonies |
</li> | </li> | ||
<li>Merike Jo ˜esaar, Eeva Heinaru, Signe Viggor, Eve Vedler & Ain Heinaru | <li>Merike Jo ˜esaar, Eeva Heinaru, Signe Viggor, Eve Vedler & Ain Heinaru | ||
| Line 63: | Line 81: | ||
<li>Jae Jun Jeong,13 Ji Hyun Kim,1 Chi-Kyung Kim,2 Ingyu Hwang3 and Kyoung Lee1 | <li>Jae Jun Jeong,13 Ji Hyun Kim,1 Chi-Kyung Kim,2 Ingyu Hwang3 and Kyoung Lee1 | ||
3- and 4-alkylphenol degradation pathway in <i>Pseudomonas</i> sp. strain KL28: genetic organization of the lap gene cluster and substrate specificities of phenol hydroxylase and catechol 2,3-dioxygenase | 3- and 4-alkylphenol degradation pathway in <i>Pseudomonas</i> sp. strain KL28: genetic organization of the lap gene cluster and substrate specificities of phenol hydroxylase and catechol 2,3-dioxygenase | ||
| + | </li> | ||
| + | <li>Passmore, I. J., Letertre, M., Preston, M. D., Bianconi, I., Harrison, M. A., Nasher, F., … Dawson, L. F. (2018). Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS pathogens, 14(9), e1007191. InterPro EMBL-EBI. “4-Hydroxy-Tetrahydrodipicolinate Synthase, DapA (IPR005263) < InterPro < EMBL-EBI.” Ebi.Ac.Uk, 2019, www.ebi.ac.uk/interpro/entry/IPR005263. Accessed 5 July 2019. | ||
</li> | </li> | ||
</html> | </html> | ||
Latest revision as of 13:50, 20 October 2019
Native promoter-pchR-sfGFP
Background
As part of our project, we decided to make a device that can sense p-Cresol. According to our research[1], we have discovered a group of bacteria known as Pseudomonas are able to sense p-Cresol naturally. In a previous study[2], the ability of Pseudomonas to sense p-Cresol is used by them to detect p-Cresol in the environment.
Part Construction & Experimental Results
The pchR-GFP was constructed from IDT DNA synthesis in the form of 3 gBlock gene fragments which contains BBa_K2997003 and BBa_K2997004 via overhang PCR. Then, we performed digestion on the finished part and ligated it into the pSB1C3 vector. Subsequently, we transformed the pchR-GFP construct into E. coli Top 10 using electroporation. After allowing the transformed E. coli Top 10 to grow on LB with Cm (Chloramphenicol) plates, colony PCR and sequencing was carried out to determine successful clones. The colony PCR results indicated that the E. coli has pSB1C3 with pchR-GFP. However, after several attempts to construct pchR-GFP, all the sequencing results indicated mutations occurred on all of our samples.

These results led us to hypothesize that the PchR protein is toxic to our E. coli. Moreover, according to a research paper[4] we have found, it is suggested in the study that p-Cresol is toxic to Gram-negative bacteria, which includes E. coli. Hence, this plasmid construct has been dropped.
Therefore, the construction of pchR-GFP was carried out again by re-doing overlap PCR, where we combine all the three IDT gene fragments into the full-length pchR-GFP construct. For this time, the finished full-length pchR-GFP part was then inserted into the vector pBBR1 MCS-4.
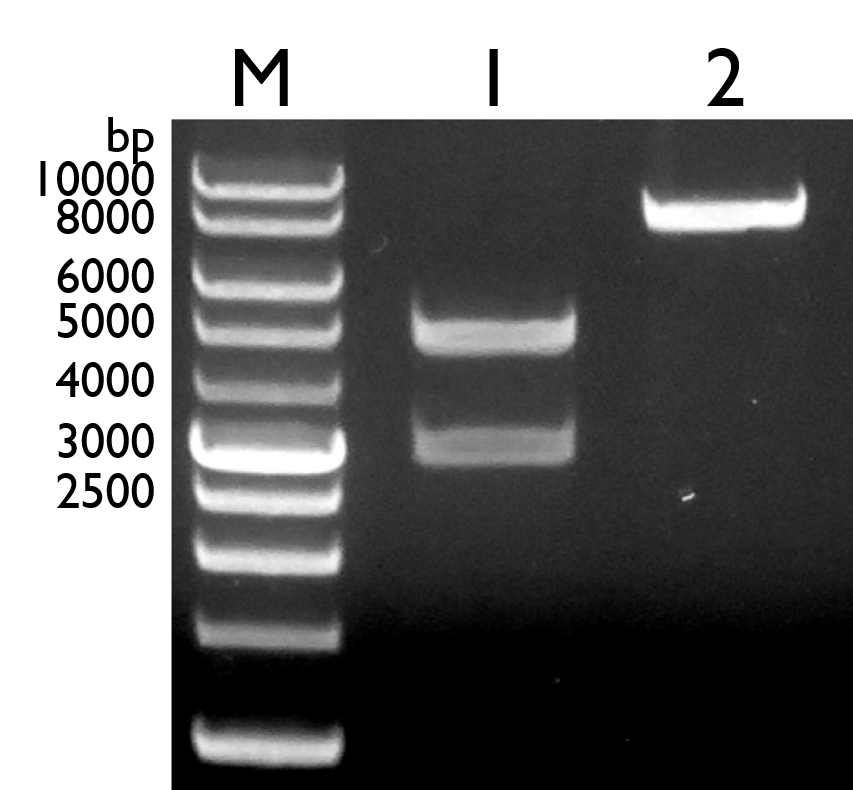
However, due to the complications surrounding the selection marker being used (Ampicillin), we were not able to get a definitive result that indicates that we have positive colonies. This is due to the fact that P.fluorescens 55 and P.aeruginosa PAD1 are naturally resistant to ampicillin and the vector we used for our construct contains ampicillin selection marker. Hence, positive colonies cannot be differentiated from negative colonies. Besides that, our colony PCR results on the single colonies were also proven to be negative.
Therefore, with time being one of the factors, we decided to drop any further construction and functional experiments surrounding the p-Cresol sensing part (pchR-GFP).
Mechanism and Biology
The construct is primarily centered upon Pseudomonas fluorescens (PC24)[2][3], as it has a gene cluster that consists of a positive inducible operon, pchR, which allows it to sense p-Cresol and promote the transcription of the downstream region to degrade the p-Cresol. The pchR gene can be translated into activator proteins that can bind with the inducer (p-Cresol) and leads to the conformation of the protein. As a result, the activator protein binds with p-Cresol and the eventual protein-p-Cresol complex will then bind onto the p-Cresol sensing region that is placed just downstream of pchR and start transcription of the gene downstream.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Illegal prefix found in sequence at 1
Illegal suffix found in sequence at 3173 - 12INCOMPATIBLE WITH RFC[12]Illegal EcoRI site found at 1
Illegal NheI site found at 1769
Illegal SpeI site found at 3174
Illegal PstI site found at 3188
Illegal NotI site found at 7
Illegal NotI site found at 3181 - 21INCOMPATIBLE WITH RFC[21]Illegal EcoRI site found at 1
Illegal BamHI site found at 1182 - 23INCOMPATIBLE WITH RFC[23]Illegal prefix found in sequence at 1
Illegal suffix found in sequence at 3174 - 25INCOMPATIBLE WITH RFC[25]Illegal prefix found in sequence at 1
Illegal XbaI site found at 16
Illegal SpeI site found at 3174
Illegal PstI site found at 3188
Illegal NgoMIV site found at 1227
Illegal AgeI site found at 210
Illegal AgeI site found at 1110 - 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI site found at 1136
Illegal BsaI.rc site found at 3096
References
- Cho, Ah Ra, Eun Jin Lim, Yaligara Veeranagouda, and Kyoung Lee* Department of Microbiology, Changwon National University, Changwon-si, Kyongnam 641-773, Korea . Identification of a p-Cresol Degradation Pathway by a GFP-Based Transposon in Pseudomonas and Its Dominant Expression in Colonies
- Merike Jo ˜esaar, Eeva Heinaru, Signe Viggor, Eve Vedler & Ain Heinaru Institute of Molecular and Cell Biology, Tartu University, Tartu, Estonia. Diversity of the transcriptional regulation of the pch gene cluster in two indigenous p-Cresol-degradative strains of Pseudomonas fluorescens
- Jae Jun Jeong,13 Ji Hyun Kim,1 Chi-Kyung Kim,2 Ingyu Hwang3 and Kyoung Lee1 3- and 4-alkylphenol degradation pathway in Pseudomonas sp. strain KL28: genetic organization of the lap gene cluster and substrate specificities of phenol hydroxylase and catechol 2,3-dioxygenase
- Passmore, I. J., Letertre, M., Preston, M. D., Bianconi, I., Harrison, M. A., Nasher, F., … Dawson, L. F. (2018). Para-cresol production by Clostridium difficile affects microbial diversity and membrane integrity of Gram-negative bacteria. PLoS pathogens, 14(9), e1007191. InterPro EMBL-EBI. “4-Hydroxy-Tetrahydrodipicolinate Synthase, DapA (IPR005263) < InterPro < EMBL-EBI.” Ebi.Ac.Uk, 2019, www.ebi.ac.uk/interpro/entry/IPR005263. Accessed 5 July 2019.
