Difference between revisions of "Part:BBa K2984006"
Chlamy Dima (Talk | contribs) |
|||
| (17 intermediate revisions by 4 users not shown) | |||
| Line 3: | Line 3: | ||
<partinfo>BBa_K2984006 short</partinfo> | <partinfo>BBa_K2984006 short</partinfo> | ||
| + | =='''Introduction'''== | ||
| + | <html> | ||
| + | For the gold medal criterium of improving an existing part, we submit this codon optimized version of the paromomycin resistance registered as <a href="https://parts.igem.org/Part:BBa_K2703008">BBa K2703008</a>. By optimizing the codon usage of this part we expected a higher expression of the antibiotic resistance enzyme and therefore a higher rate of success when using this resistance gene for paromomycin resistance. Our characterization resulted in a confirmation of our hypothesis and a successful improvement of the BBa_K2703008 part. | ||
| + | </html> | ||
| − | |||
===Usage and Biology=== | ===Usage and Biology=== | ||
| + | |||
| + | Paromomycin belongs to a group of aminoglycoside antibiotics such as neomycin or dibekacin. These aminoglycosides are capable of inhibiting the eukaryotic translation, by binding within the large and small subunit of the 80S ribosome. This property allows paromomycin to be used as a selection marker. The paromomycin resistance BBa_K2703008, registered by the Cambridge team 2016, works by allowing the expression of an enzyme which catalyses the transfer of the gamma-phosphate of ATP to the hydroxyl group in 3’ position of the paromomycin molecule and allows the carrier of the gene to develop a resistance to paromomycin. Some organisms exhibit a higher expression of transgene enzymes when the genetic code is codon optimized for the specific organism. One of these organisms is the alga <i>Chlamydomonas reinhardtii</i>. With the following part, we propose a codon optimized version of the paromomycin resistance for use in <i>Chlamydomonas reinhardtii</i>. This part is optimal for paromomycin screening when using <i>C. reinhardtii</i>. | ||
<!-- --> | <!-- --> | ||
| Line 11: | Line 16: | ||
<partinfo>BBa_K2984006 SequenceAndFeatures</partinfo> | <partinfo>BBa_K2984006 SequenceAndFeatures</partinfo> | ||
| − | ==''' | + | =='''Characterization'''== |
| − | + | <html> | |
| + | <h3> Methods </h3> | ||
| + | |||
| + | To see if the expression of the aminoglycoside 3’-phosphotransferase was increased, we performed several electroporations to transform <i>C. reinhardtii</i> with the improved paromomycin resistance. We used the <i>C. reinhardtii</i> strain UVM 4 since it is a strain designed to express transgene constructs (Neupert et al. 2009). We transformed the two paromomycin constructs with standard and improved codon usage starting with 0,5 µg DNA per electroporation sample and ascended with 0,5 µg steps up to 2 µg. For each construct and DNA mass we did three electroporations. The electroporation electrical resistance was measured for each sample. After resuspension and one day recovery in TAP medium, all samples were plated on TAP-agar plates containing a paromomycin concentration of 10 µM. After two weeks of growth, colonies corresponding to each sample were counted. Each colony of <i>C.reinhardtii</i> represents a successful transformation of the resistance and indicates the expression of the aminoglycoside 3’-phosphotransferase. By counting the amount of colonies on the plates, we could determine which construct and at which DNA mass at the time of transformation worked best. | ||
| + | |||
| + | <h3> Results </h3> | ||
| + | |||
| + | First, we counted the total number of <i>C. reinhardtii</i> colonies that were transformed with the standard and improved resistance, regardless of the DNA mass used at the time of transformation. The results of the total colony count can be seen in Fig. 1. Counting the colonies we discovered that the total number of colonies was much higher for the improved plasmid version. The colonies of the samples using the standard usage resulted in a total amount of 175 whereas the improved plasmid version produced 665 colonies. This result indicates that the improved paromomycin resistance works better than the one with standard codon usage. | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://2019.igem.org/wiki/images/a/a7/T--Humboldt_Berlin--Colony_Amount_Total.png" alt="colonies_total" width="500"> | ||
| + | <figcaption>Fig.1 - Total amount of colonies counted for the standard (blue) and improved (orange) resistance.</figcaption> | ||
| + | </figure> | ||
| + | |||
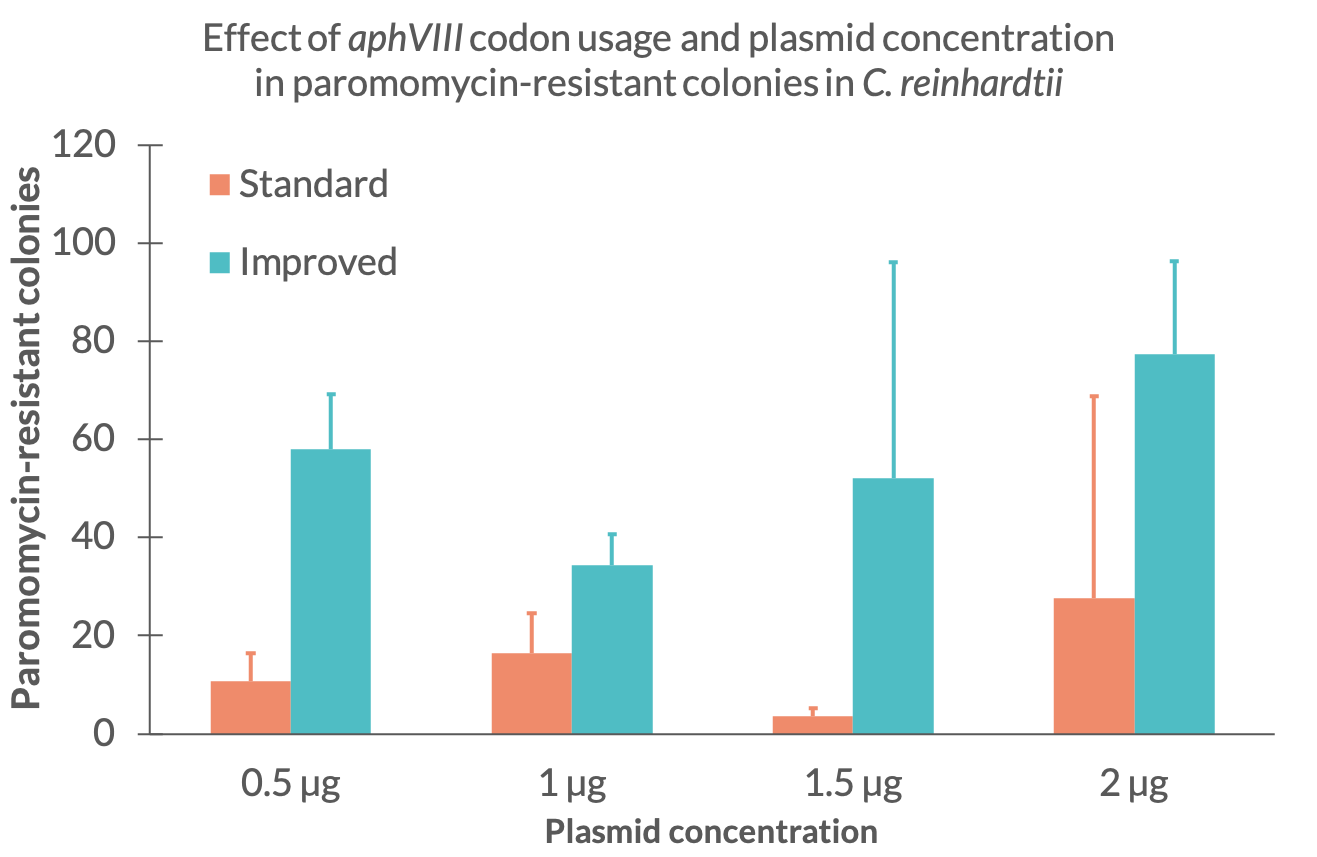
| + | Comparing the plasmid mass at the time of electroporation we discovered that the amount of colonies does not strictly correlate to the amount of DNA used during the electroporation (Fig. 2.). For the paromomycin resistance with standard codon usage we can see that the number of colonies at 1,5 µg is smaller than expected. Similarly, the amount of colonies for the improved resistance at a DNA mass of 0,5 µg is much higher than expected. The other results seem to show a tendency of increasing colony numbers with more DNA mass. Yet, further tests should be made to examine the exact effect of DNA mass during transformation for these parts. | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://2019.igem.org/wiki/images/9/9d/T--Humboldt_Berlin--Colony_Amount_Mean_3.png" alt="colonies_mean" width="600"> | ||
| + | <figcaption>Fig 2. - Mean colony amount for the standard (blue) and improved (orange) paromomycin resistance for different DNA mass at the time of electroporation</figcaption> | ||
| + | </figure> | ||
| + | |||
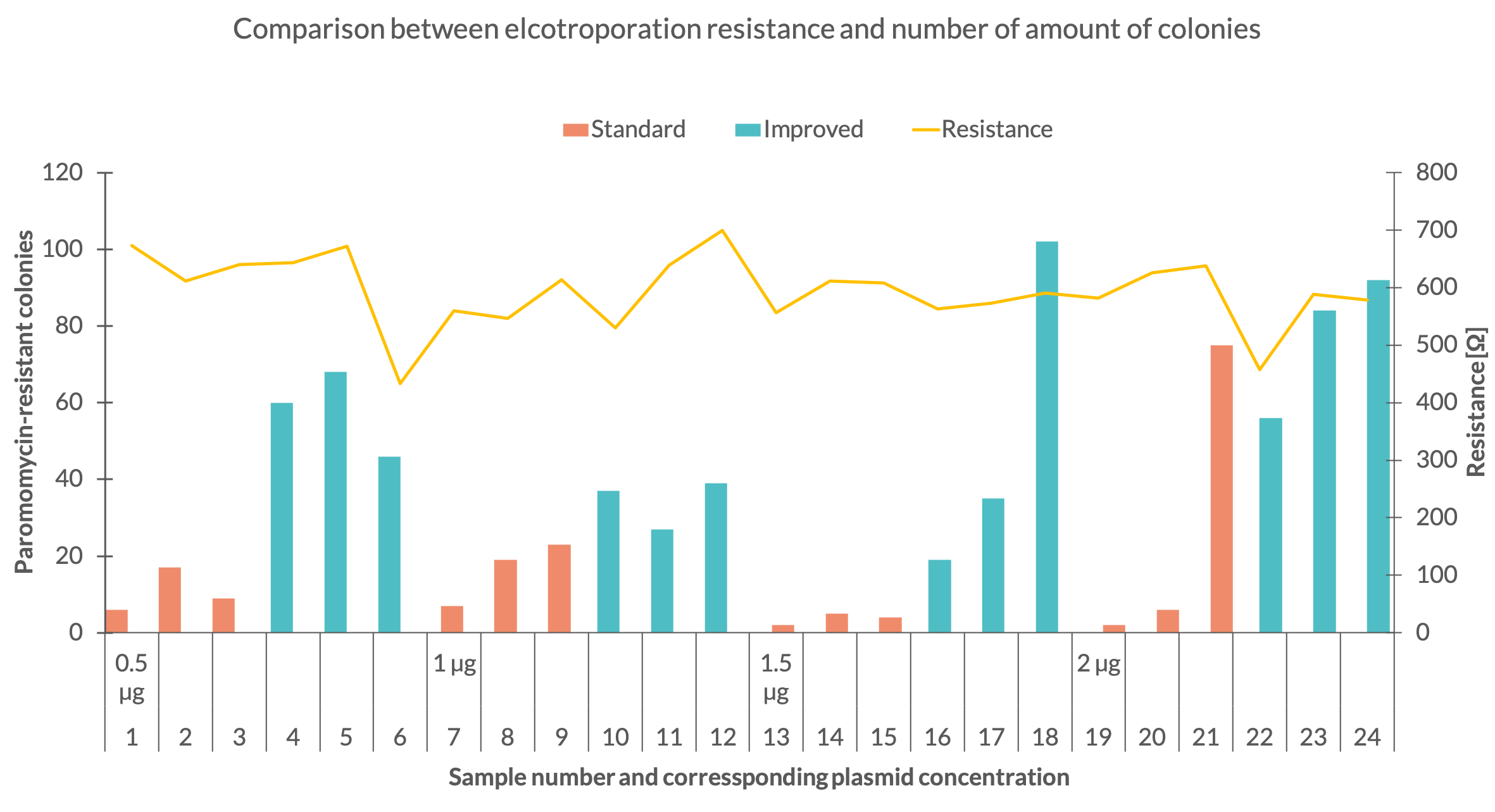
| + | One explanation for the variable amount of colonies might be the inconsistency of the electroporation resistance. To see how the electroporation process affected the number of colonies their quantity was compared with the corresponding resistance (Fig. 3). Fig. 3 depicts that the set with 1,5 µg standard plasmid was executed with a robust resistance of around 570 Ω for all 3 samples. The 1 µg set of the same plasmid shows a variable resistance but delivered more colonies. In the 2 µg standard plasmid set the resistance of the first sample dropped to 457 Ω but the same amount of colonies as in sample 1 of the 1,5 µg standard set were counted. With further comparison of these to sets it can be seen, that the third samples in the 1,5 µg and 2 µg sets showed similar resistance but the third sample of the second set resulted in a much higher colony number. The data behaves similar for the improved plasmid. The second sample of the first set and the third sample of the third were carried out with a resistance around 610 Ω but for the third sample almost 34 more colonies were counted. | ||
| + | |||
| + | <figure> | ||
| + | <img src="https://2019.igem.org/wiki/images/c/c8/T--Humboldt_Berlin--Colonies_with_resistance.png" alt="colonies_resistance" width="700"> | ||
| + | <figcaption>Fig 3. - Amount of colonies for each sample in dependency of the DNA mass and plotted with the electrical resistance of the electroporation</figcaption> | ||
| + | </figure> | ||
| + | |||
| + | The experiments showed clearly, that the improved codon usage resulted in larger number of <i>C.reinhardtii</i> colonies capable of growing TAP-paromomycin-agar plates. This proves that the changed codon usage promotes a higher expression of the aminoglycoside 3’-phosphotransferase and is therefore an improvement of part BBa_K2703008. | ||
| + | |||
| + | |||
| + | </html> | ||
| + | =='''References'''== | ||
| + | [1] Neupert, J., Karcher, D., & Bock, R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. The Plant Journal, 57(6), 1140-1150. | ||
| − | < | + | [2] Sizova, I., Fuhrmann, M., & Hegemann, P. (2001). A Streptomyces rimosusaphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to <i>Chlamydomonas reinhardtii</i>. Gene, 277(1-2), 221-229. |
| − | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 13:46, 20 October 2019
Codon-Usage-Optimized Paromomycin Resistance For Use in Chlamydomonas reinhardtii
Introduction
For the gold medal criterium of improving an existing part, we submit this codon optimized version of the paromomycin resistance registered as BBa K2703008. By optimizing the codon usage of this part we expected a higher expression of the antibiotic resistance enzyme and therefore a higher rate of success when using this resistance gene for paromomycin resistance. Our characterization resulted in a confirmation of our hypothesis and a successful improvement of the BBa_K2703008 part.
Usage and Biology
Paromomycin belongs to a group of aminoglycoside antibiotics such as neomycin or dibekacin. These aminoglycosides are capable of inhibiting the eukaryotic translation, by binding within the large and small subunit of the 80S ribosome. This property allows paromomycin to be used as a selection marker. The paromomycin resistance BBa_K2703008, registered by the Cambridge team 2016, works by allowing the expression of an enzyme which catalyses the transfer of the gamma-phosphate of ATP to the hydroxyl group in 3’ position of the paromomycin molecule and allows the carrier of the gene to develop a resistance to paromomycin. Some organisms exhibit a higher expression of transgene enzymes when the genetic code is codon optimized for the specific organism. One of these organisms is the alga Chlamydomonas reinhardtii. With the following part, we propose a codon optimized version of the paromomycin resistance for use in Chlamydomonas reinhardtii. This part is optimal for paromomycin screening when using C. reinhardtii.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 298
Illegal NotI site found at 122 - 21INCOMPATIBLE WITH RFC[21]Illegal XhoI site found at 13
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 401
- 1000COMPATIBLE WITH RFC[1000]
Characterization
Methods
To see if the expression of the aminoglycoside 3’-phosphotransferase was increased, we performed several electroporations to transform C. reinhardtii with the improved paromomycin resistance. We used the C. reinhardtii strain UVM 4 since it is a strain designed to express transgene constructs (Neupert et al. 2009). We transformed the two paromomycin constructs with standard and improved codon usage starting with 0,5 µg DNA per electroporation sample and ascended with 0,5 µg steps up to 2 µg. For each construct and DNA mass we did three electroporations. The electroporation electrical resistance was measured for each sample. After resuspension and one day recovery in TAP medium, all samples were plated on TAP-agar plates containing a paromomycin concentration of 10 µM. After two weeks of growth, colonies corresponding to each sample were counted. Each colony of C.reinhardtii represents a successful transformation of the resistance and indicates the expression of the aminoglycoside 3’-phosphotransferase. By counting the amount of colonies on the plates, we could determine which construct and at which DNA mass at the time of transformation worked best.Results
First, we counted the total number of C. reinhardtii colonies that were transformed with the standard and improved resistance, regardless of the DNA mass used at the time of transformation. The results of the total colony count can be seen in Fig. 1. Counting the colonies we discovered that the total number of colonies was much higher for the improved plasmid version. The colonies of the samples using the standard usage resulted in a total amount of 175 whereas the improved plasmid version produced 665 colonies. This result indicates that the improved paromomycin resistance works better than the one with standard codon usage.
References
[1] Neupert, J., Karcher, D., & Bock, R. (2009). Generation of Chlamydomonas strains that efficiently express nuclear transgenes. The Plant Journal, 57(6), 1140-1150.
[2] Sizova, I., Fuhrmann, M., & Hegemann, P. (2001). A Streptomyces rimosusaphVIII gene coding for a new type phosphotransferase provides stable antibiotic resistance to Chlamydomonas reinhardtii. Gene, 277(1-2), 221-229.
