Difference between revisions of "Part:BBa K3183200"
| Line 5: | Line 5: | ||
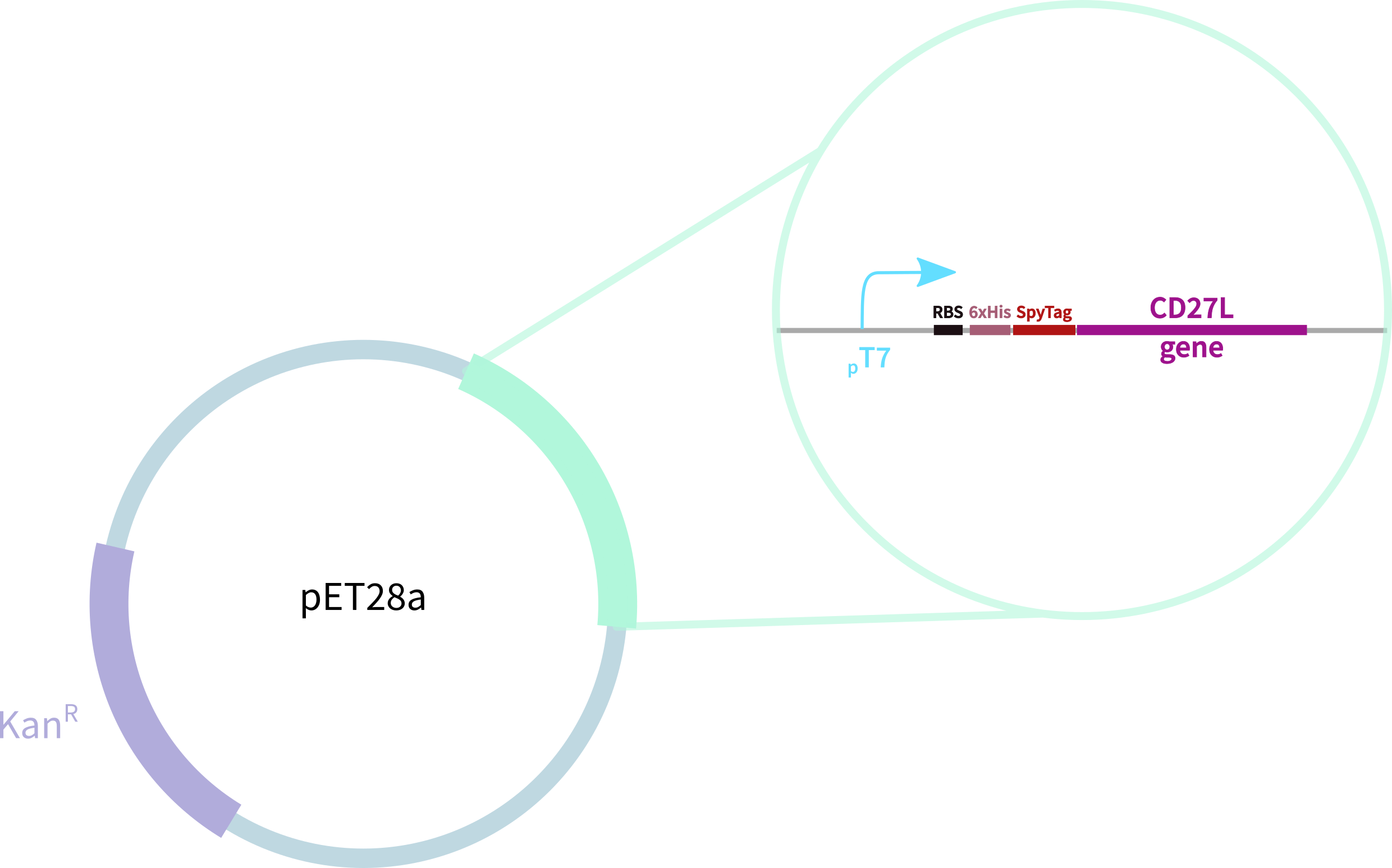
This part contains CD27L fused to an N-Terminal 6-His Tag and SpyTag for purification, oligomerisation, and other SpyCatcher applications. This endolysin has previously been shown to be specific to <i>C. difficile</i>. This endolysin works by cleaving residues<sup>1</sup> (Fig. 1), breaking the cell's peptidoglycan and compromising the structural integrity of its cell wall. Two X-ray crystal structures were solved for the N-terminal<sup>2</sup> and C-terminal domains<sup>3</sup>. These are joined by a linker, which we've added ourselves (Figure 2). | This part contains CD27L fused to an N-Terminal 6-His Tag and SpyTag for purification, oligomerisation, and other SpyCatcher applications. This endolysin has previously been shown to be specific to <i>C. difficile</i>. This endolysin works by cleaving residues<sup>1</sup> (Fig. 1), breaking the cell's peptidoglycan and compromising the structural integrity of its cell wall. Two X-ray crystal structures were solved for the N-terminal<sup>2</sup> and C-terminal domains<sup>3</sup>. These are joined by a linker, which we've added ourselves (Figure 2). | ||
| − | [[File:BBa_K3183009_Structure.png|thumb|right|430px|'''Figure 2:''' X-ray Crystal Structure of CD27L; PDB codes 3QAY (green) & 4CU5 ( | + | [[File:BBa_K3183009_Structure.png|thumb|right|430px|'''Figure 2:''' X-ray Crystal Structure of CD27L; PDB codes 3QAY (green) & 4CU5 (purple) with a linker (yellow) drawn in with PyMol by Team Oxford 2019.]] |
<br><br><br><br> | <br><br><br><br> | ||
[[File:BBa_K3183009_Reaction.png|thumb|left|430px|'''Figure 1:''' CD27L Catalytic Reaction.]] | [[File:BBa_K3183009_Reaction.png|thumb|left|430px|'''Figure 1:''' CD27L Catalytic Reaction.]] | ||
Revision as of 11:21, 20 October 2019
CD27L with N-Terminal 6-His Tag and SpyTag
This part contains CD27L fused to an N-Terminal 6-His Tag and SpyTag for purification, oligomerisation, and other SpyCatcher applications. This endolysin has previously been shown to be specific to C. difficile. This endolysin works by cleaving residues1 (Fig. 1), breaking the cell's peptidoglycan and compromising the structural integrity of its cell wall. Two X-ray crystal structures were solved for the N-terminal2 and C-terminal domains3. These are joined by a linker, which we've added ourselves (Figure 2).
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Characterisation
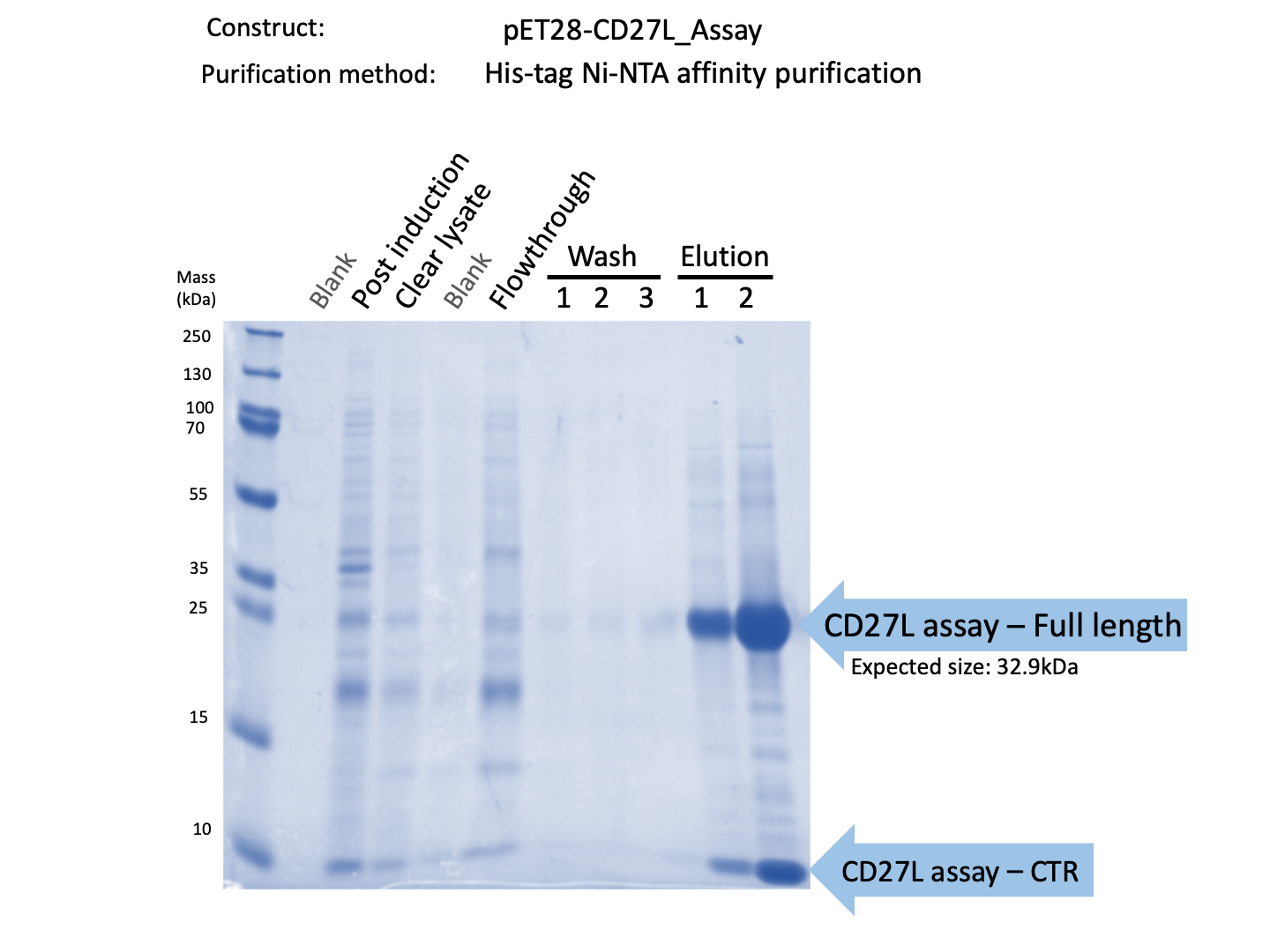
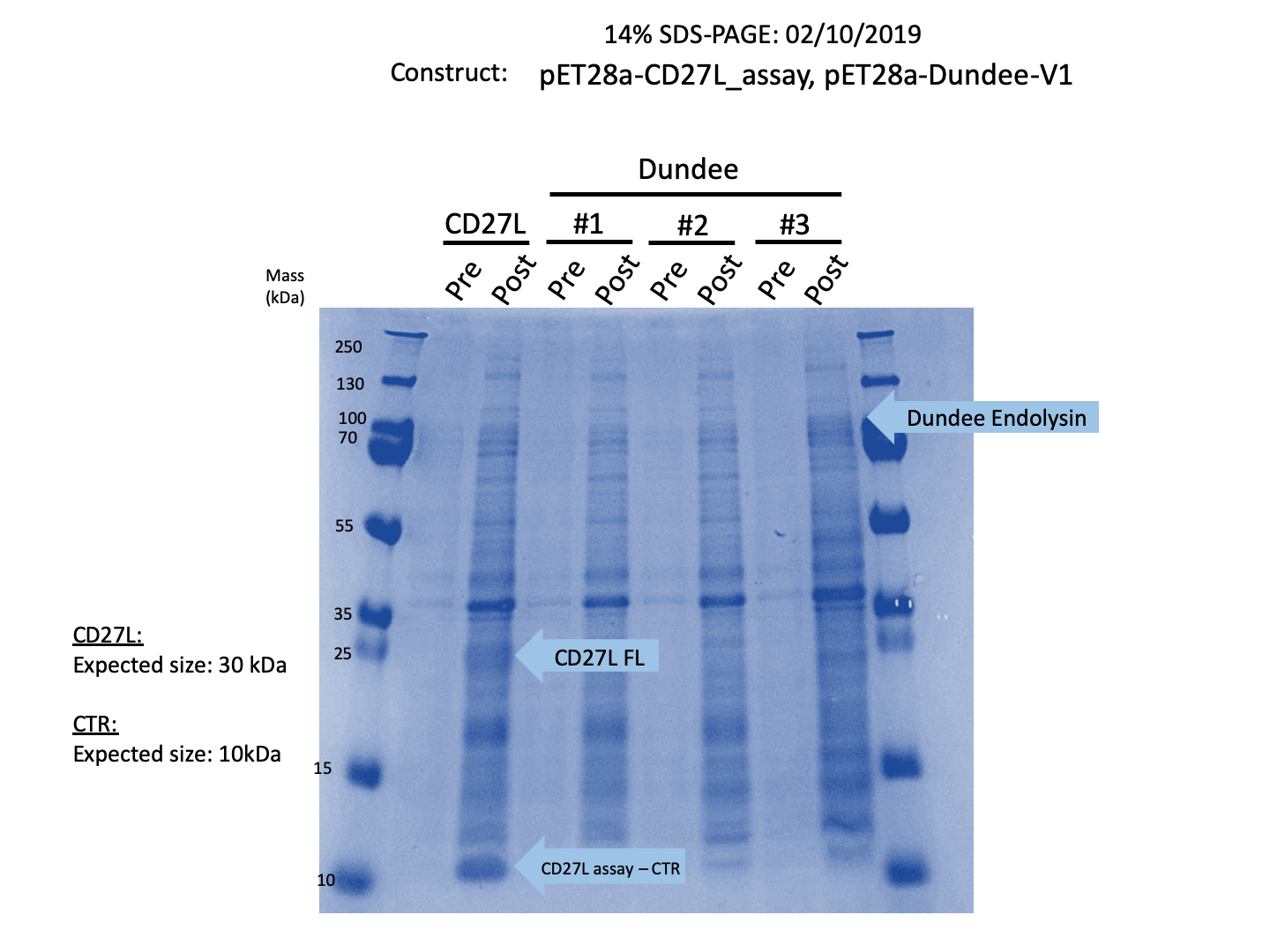
We expressed endolysin in the pET28A vector following difficulties with Dundee's CD27L1-179 endolysin BBa_K895005. We were able to achieve strong expression under the pET28A system (Fig. 3).
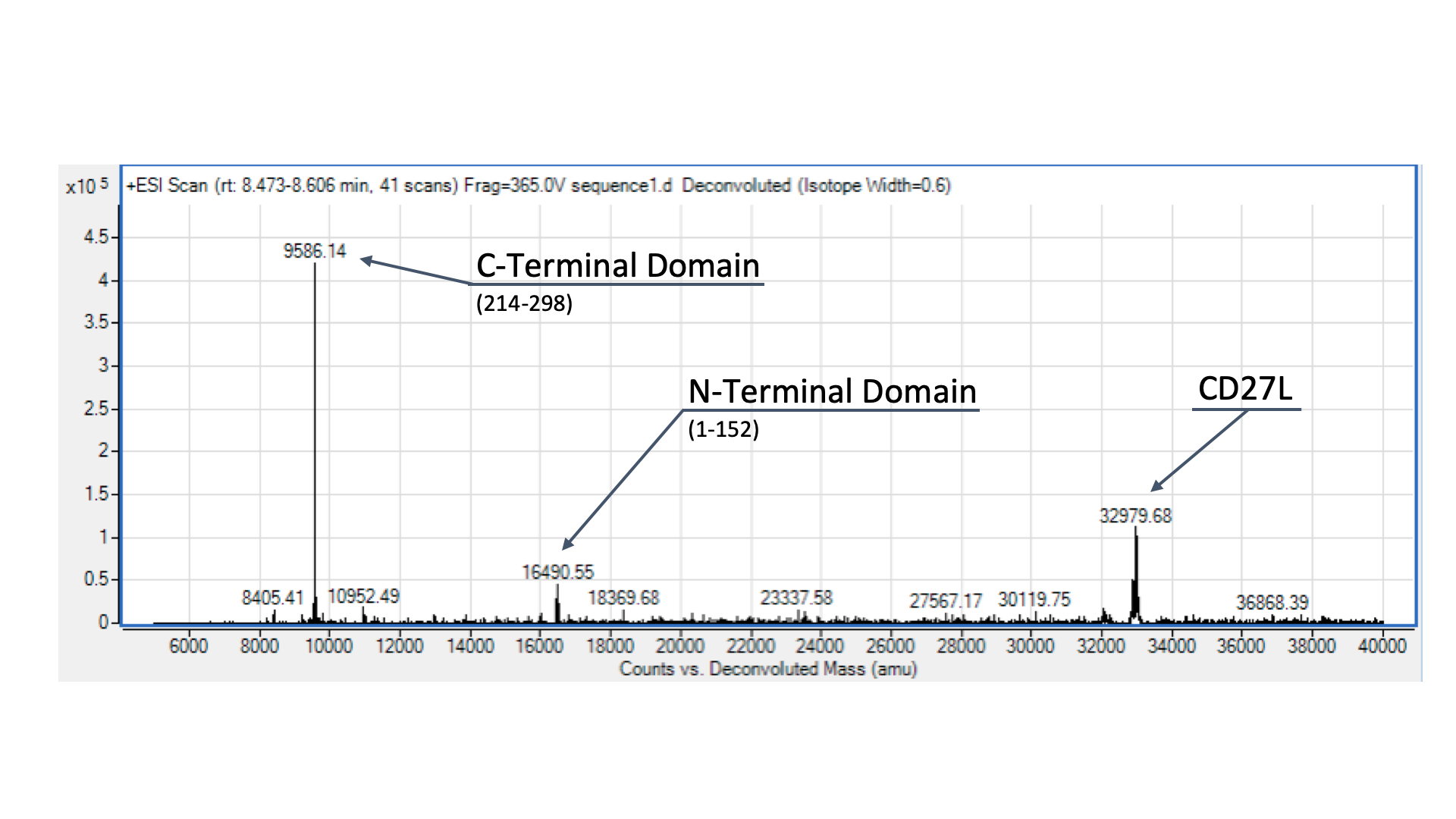
To confirm the identity of our endolysin, we wanted to run it through electrospray mass spectrometry (ESI-MS). However, given that we didn’t have access to size-exclusion chromatography, we were only able to send our CD27L protein preparation, as the other sample was far too dirty for effective ESI-MS. The identity of our endolysin, as well as putative autocleavage is shown and discussed in Fig. 4.
Due to safety concerns, our endolysin killing assays could only be carried out on on Bacillus subitlis, a Category 1 bacteria as opposed to Clostridium difficile, a Category 2 bacteria. B. subtilis has a similar composition of peptidogylcan cell wall to C. difficile2, allowing us to quantify killing data.
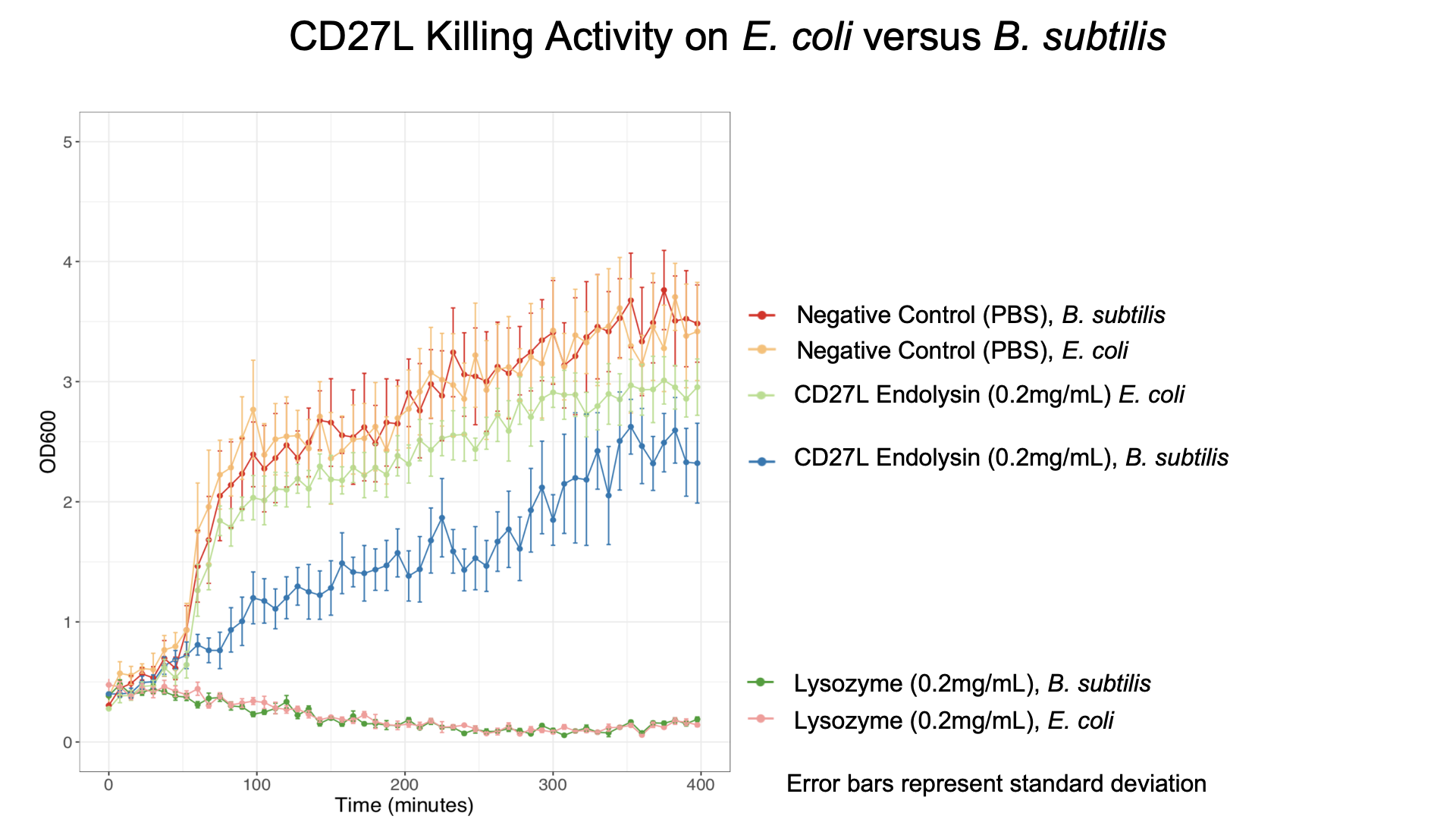
To get more information about the killing kinetics of the CD27L endolysin, we titrated our killing assay with increasing concentrations of CD27L endolysin. This titration showed measurable effect on B. subtilis cultures in LB broth with final endolysin concentrations greater than 0.2mg/mL (Fig. 5). Rate constants regarding how much the endolysin inhibited growth were subsequently used to inform our model (Table 2).
From this data, we were able to obtain kinetic constants for endolysin killing, which were subsequently used in our mathematical models.

We also showed that endolysin was specific to B. subtilis using a killing assay involving E. coli, B. subtilis, and our chassis Lactobacillus reuteri (Fig. 6 & 7).

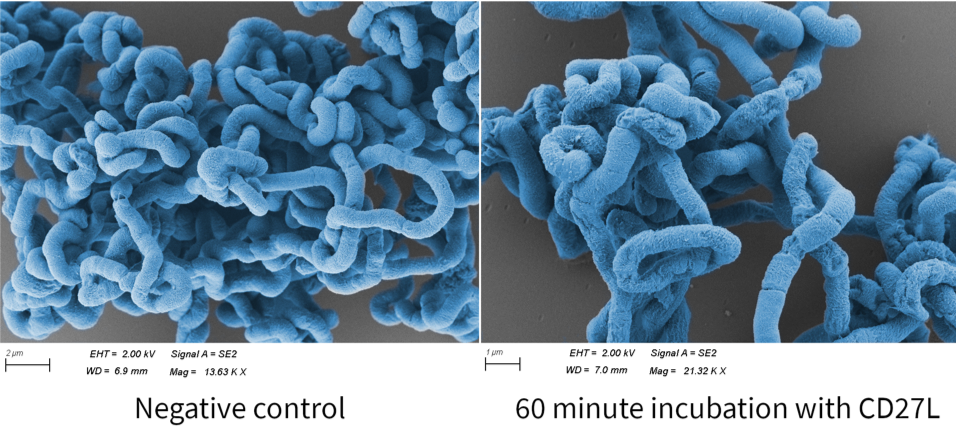
Finally, we fortunate enough to obtain scanning electron microscopy (SEM) images thanks to Errin Johnson at the Dunn School of Pathology. After performing our endolysin killing assay, we fixed the cells onto glass slides and imaged them. We chose this as a qualitative method to determine if CD27L had a similar effect on endolysin killing as lysozyme4. Below shows images of B. subtilis following CD27L endolysin treatment (Fig. 8). Here, we demonstrated some changes in B. subtilis cell morphology, but further investigation is needed to explore these effects.
Part Improvement of BBa_K895005
A fundamental crux of the ProQuorum system is the expression of the endolysin which is responsible for the actual lysis and consequent killing of the pathogenic C. difficile. Thus, there is a clear need for an endolysin that fulfills the criteria of being:
1. optimally expressed
2. optimally secreted and
3. optimally effective against C.difficile
Thus, we redesigned the BBa_K895005 part submitted by the 2012 Dundee iGEM team, which consists of:
1. truncated form of the CD27L endolysin, shown to increase both efficacy and host range2
2. Type VI secretion protein derived from S. typhimurium
3. Hemagglutinin (HA) Tag for Western Blotting</b>
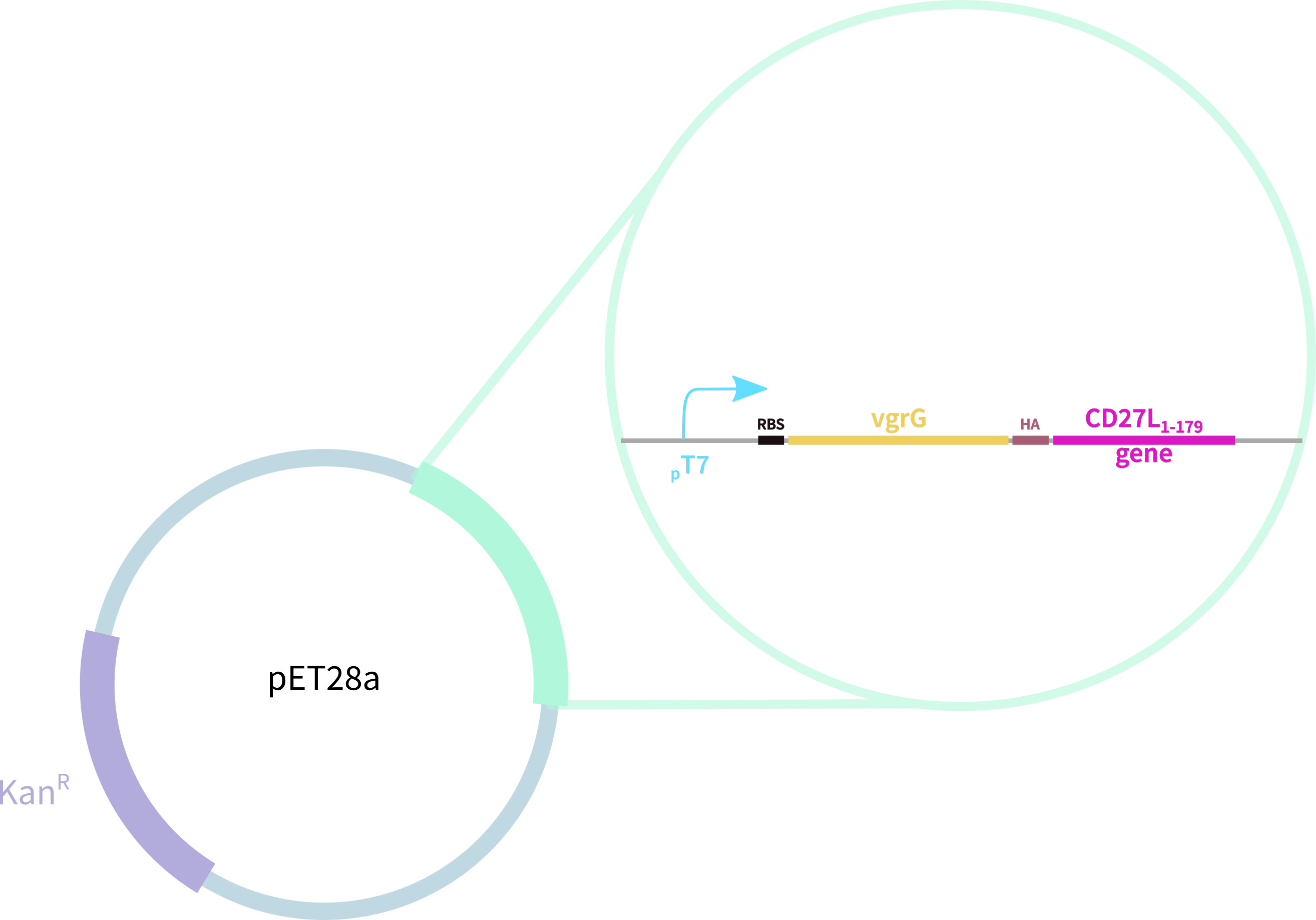
In contrast, our novel part BBa_K3183200 (CD27L) encodes:
1. full-length form of the CD27L endolysin
2. SpyTag for purification, oligomerisation, and other SpyCatcher applications
3. 6-His tag for easy purification
Incorporating both of these parts into pET28A, our expression vector, we planned on testing the relative efficacies of B.subtilis killing as per our killing assays. Thus, we followed our generic pipeline of:
1. transformation of both constructs into E.coli (BL21 (DE3)-RIPL Competent E.coli)
2. miniprep + sequencing to verify successful transformation
3. induction of expression with IPTG
Thus, we sought out the endolysin in an existing part made by the 2012 iGEM Team Dundee. However, despite identical growth and induction conditions, expression of the Dundee 2012 iGEM endolysin was unsuccessful, even in triplicate, as shown in Figure 2. In contrast, expression of the CD27L_Assay part was successful and quantitatively verified via both mass spectrometry and BCA assay, as in the Results page.
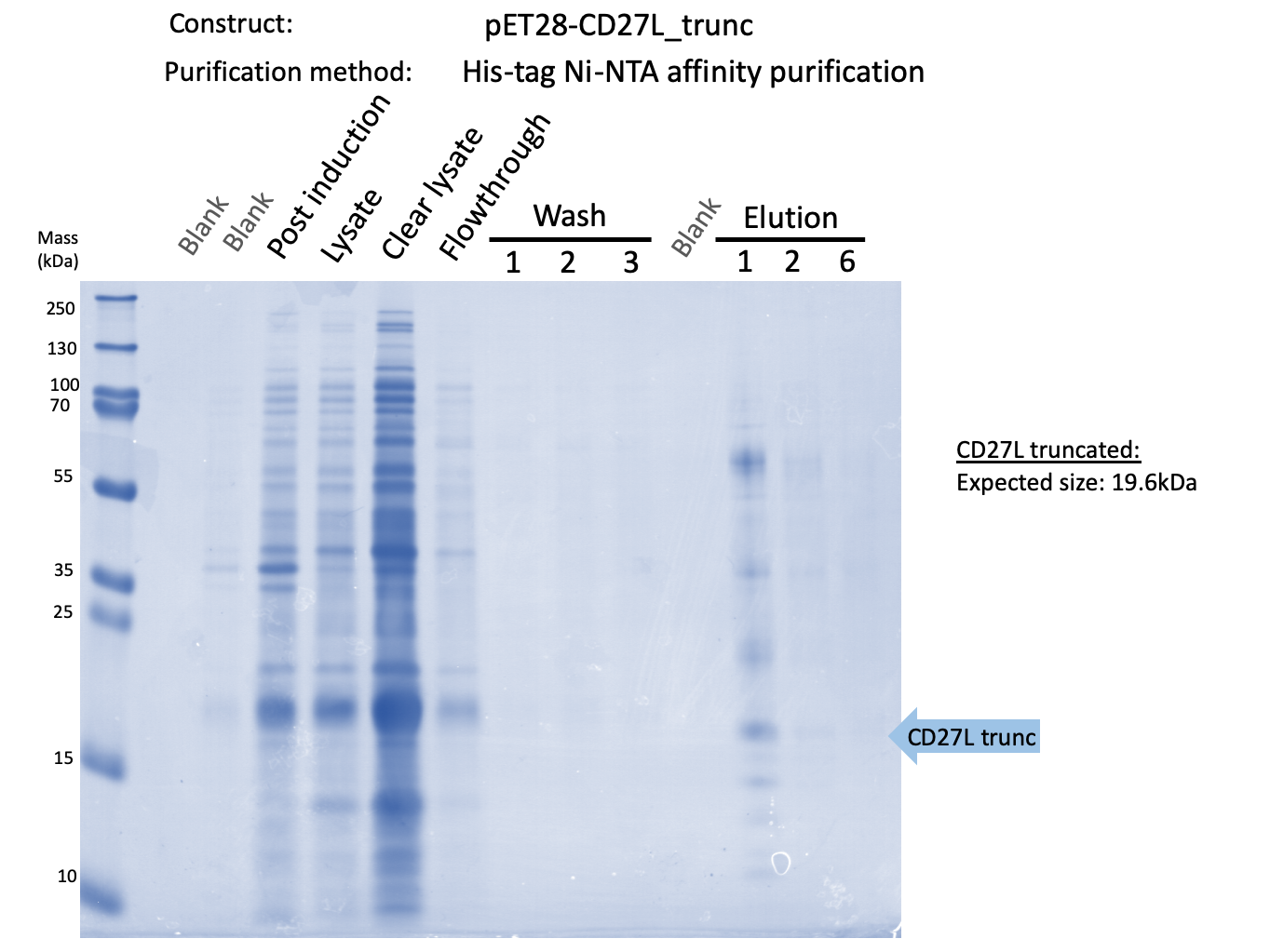
Thus, to solve this issue, we decided to attempt expression of only the truncated endolysin from the Dundee 2012 Biobrick, without its substantially large Type VI secretion tag. We added 6His and SpyTag to the N-terminus to allow for detection (via SpyCatcher gel-shift assays) and purification. This resulted in BBa_K31830201 (CD27L1-179). Upon analysis, there is evident expression of the truncated endolysin as seen in Figure 3.1. However, given identical conditions of growth and induction, there appears to be greater expression of the full-length endolysin (Fig. 3.2), which may perhaps indicate greater stability.
As a further improvement, we decided to measure the killing efficacy of the CD27L_Assay endolysin on B. subtilis (our surrogate target) as no killing data was provided for the Dundee 2012 BBa_K895005 part. As seen in Figure 4, there is a substantial decrease in OD600 over time relative to the negative controls.
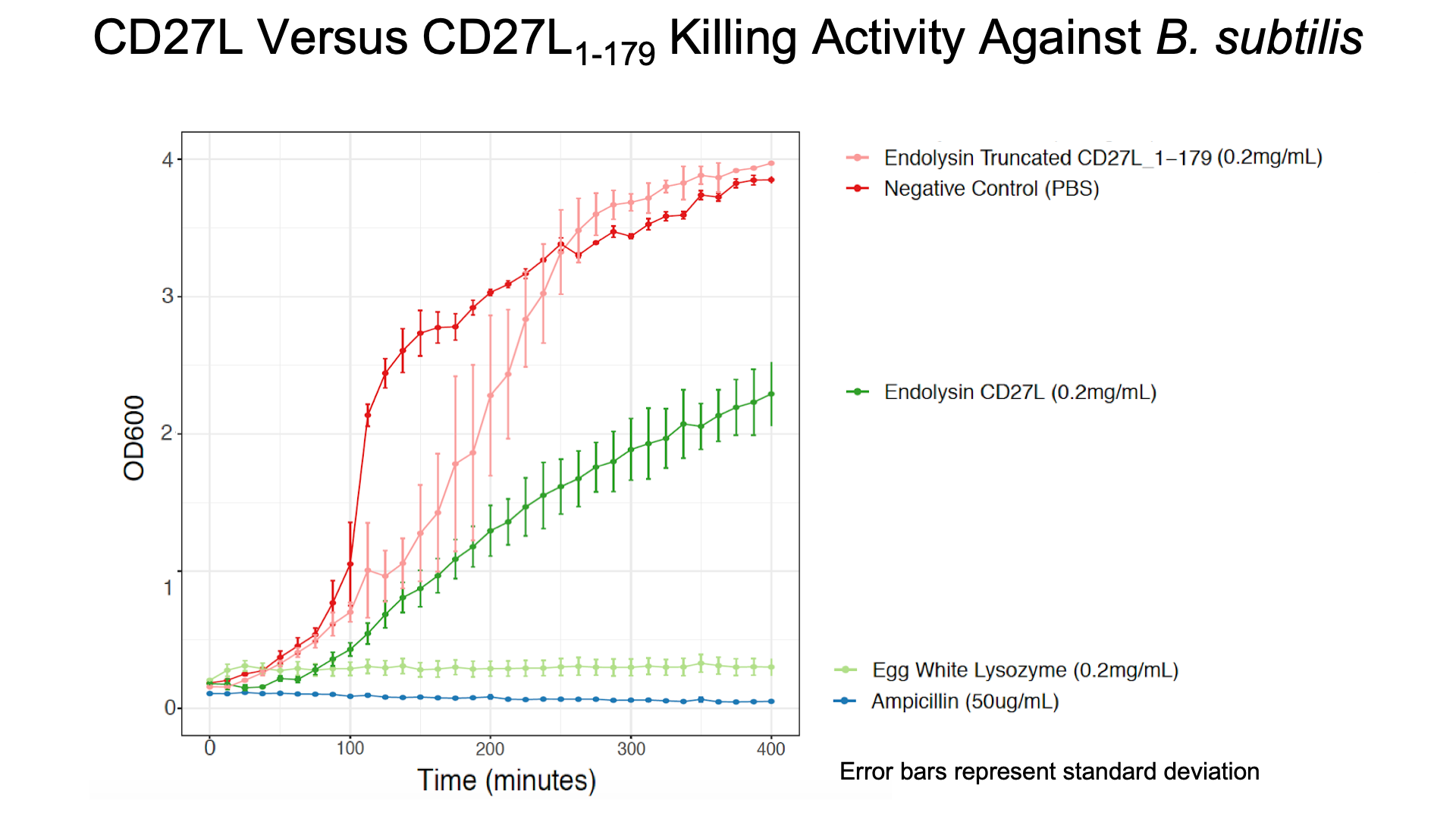
The truncated CD27L1-179 shows decreased growth relative to the negative control; however, log-phase growth resumes after 150 minutes. CD27L endolysin results in decreased growth during the first 100 minutes, and growth thereafter appears to be linear. This may point to inhibited log-phase growth in subsequent generations of B. subtilis following CD27L exposure.
References
1. Mayer, M. J., et al. “Molecular Characterization of a Clostridium Difficile Bacteriophage and Its Cloned Biologically Active Endolysin.” Journal of Bacteriology, vol. 190, no. 20, 2008, pp. 6734–6740., doi:10.1128/jb.00686-08.
2. Mayer, M. J et al. “Structure-based modification of a Clostridium difficile-targeting endolysin affects activity and host range.” Journal of bacteriology vol. 193,19 (2011): 5477-86. doi:10.1128/JB.00439-11
3. Dunne, Matthew et al. “The CD27L and CTP1L endolysins targeting Clostridia contain a built-in trigger and release factor.” PLoS pathogens vol. 10,7 e1004228. 24 Jul. 2014, doi:10.1371/journal.ppat.1004228
4. Twetman, Svante, et al. “Scanning Electron Microscopic Study of Streptococcus Mutans BHT Lysed by Lysozyme.” European Journal of Oral Sciences, vol. 93, no. 1, 1985, pp. 23–29., doi:10.1111/j.1600-0722.1985.tb01304.x.