Difference between revisions of "Part:BBa K3254011"
Zongyeqing (Talk | contribs) |
Zongyeqing (Talk | contribs) (→Results) |
||
| Line 30: | Line 30: | ||
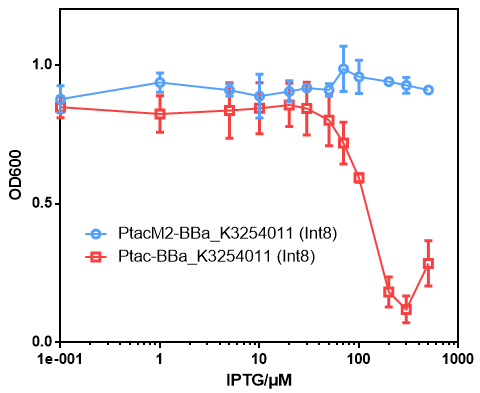
*We attempt to use a mutant Ptac promoter([[Part:BBa_K3254016|Ptac-M2]] to solve the problems of strong leakage and toxicity when this part was transcripted by [[Part:BBa_K2572025|Ptac]]. | *We attempt to use a mutant Ptac promoter([[Part:BBa_K3254016|Ptac-M2]] to solve the problems of strong leakage and toxicity when this part was transcripted by [[Part:BBa_K2572025|Ptac]]. | ||
| − | *No significant toxicity was observed during induction when using the Ptac-M2 promoter, however, the leakage expression is still strong which might due to the strong RBS. | + | *No significant toxicity was observed during induction when using the Ptac-M2 promoter, however, the leakage expression is still strong which might due to the strong RBS (Data not shown). |
[[File:T--GENAS_China--IPTG-int8OD600.png]] | [[File:T--GENAS_China--IPTG-int8OD600.png]] | ||
| − | |||
=Orthogonality Characterization= | =Orthogonality Characterization= | ||
Revision as of 09:37, 20 October 2019
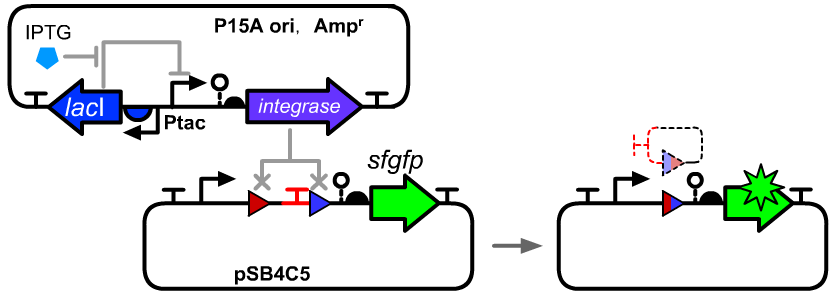
Translational Unit for INT8 integrase
This part can be placed downstreamed a prmoter. The leakage transcription of a Ptac promoter on a P15A ori plasmid was sufficient for the recombination of the att sites on low copy vectors.
- We characterized the toxicity of this part, and the orthogonality between Int8 and 5 other LSTP using this part.
Toxicity Characterization
- The cis-element used in this experiments were BBa_K3254003 which contains the Int10 att sites.
- We constructed an IPTG inducible system to control the expressional level by the IPTG concentration in E.coli DH5α host.
Genetic Design
- The composition and principle of the experimental system are indicated below. More details can be seen on the pages of BBa_K3254003.
- This part was placed downstream of a Ptac promoter.BBa_K3254025 can be seen as a reference of device structure.
Experimental Setup
The two plasmids were co-transformed into E.coli DH5α host. Then we selected colonies with no observable fluoresce and inoculated into M9 supplemented medium for overnight growth. Then, the cell cultures were diluted 1000-fold with M9 supplemented medium with gradient concentrations of IPTG inducer and growth for another 20 hours. All incubations were carried out using a Digital Thermostatic Shaker maintained at 37 °C and 1000 rpm, using flat-bottom 96-well plates sealed with sealing film. Finally, the OD600 of each well was measured by a microplate reader.
- The differents between the readings of OD600 of each group could be a reference of toxicity characteristic.
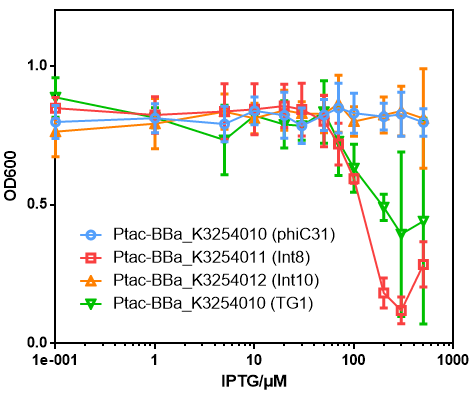
Results
- The data points with red color is the results of this part.
- Compared to other groups, Significant toxicity was observed when induced by high-concentration IPTG.
- We attempt to use a mutant Ptac promoter(Ptac-M2 to solve the problems of strong leakage and toxicity when this part was transcripted by Ptac.
- No significant toxicity was observed during induction when using the Ptac-M2 promoter, however, the leakage expression is still strong which might due to the strong RBS (Data not shown).
Orthogonality Characterization
We conducted orthogonality tests to see the compatibility between the 6 integrase used in our project. The other integrases include phiC31, Int5, Int7, Int10 and TG1.
Genetic Design
Similar to the design of thermodynamic characterization. Each integrase generator plasmid were co-transferred with 6 plasmid containing different sets of cis-elements.
Experimental Setup
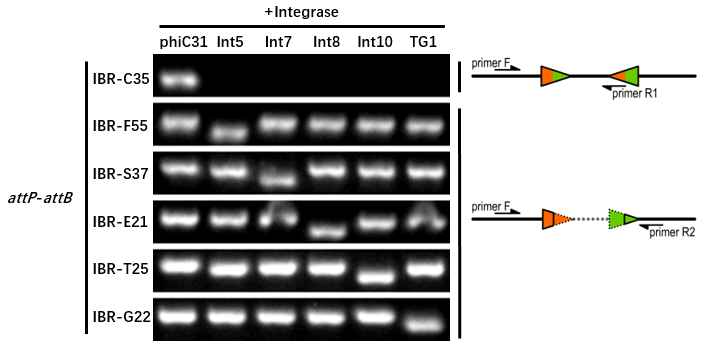
The growth condition is similar to the thermodynamic characterization but the other samples were full induced with 500 μM IPTG. Then we used specific primers to identify the genotype of att sites by PCR. The principle of genotype identification was shown on the right of result image.
Results
The results indicate that Int8 integrase has a good orthogonality with the other 5 att sites and compatible with other integrases. IBR-C35/F55/S37/E21/T25/G22 were the plasmids with phiC31/Int5/Int7/Int8/Int10/TG1 att sites.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BglII site found at 1327
- 23COMPATIBLE WITH RFC[23]
- 25INCOMPATIBLE WITH RFC[25]Illegal AgeI site found at 1030
Illegal AgeI site found at 1427 - 1000COMPATIBLE WITH RFC[1000]