Difference between revisions of "Part:BBa K3196014"
GlacierHOLE (Talk | contribs) |
GlacierHOLE (Talk | contribs) |
||
| Line 10: | Line 10: | ||
<h1>'''DNA Gel Electrophoretic'''</h1> | <h1>'''DNA Gel Electrophoretic'''</h1> | ||
After we link PHO1 αpro and SLAC successfully, we run the PCR with an intention to confirm the expression of SLAC. As the figure shows, we get the genetic stripe about 2019 bp which means the PCR is successful. | After we link PHO1 αpro and SLAC successfully, we run the PCR with an intention to confirm the expression of SLAC. As the figure shows, we get the genetic stripe about 2019 bp which means the PCR is successful. | ||
| − | [[File: | + | [[File:T--HUST-China--2019-DNA Gel Electrophoretic.png|400px|thumb|center|Figure1:This is the result of the SacⅠ single endonuclease digestion plasmid of the TOP10 strain, we will extract the gel and obtain the aimed DNA. ]] |
| + | |||
<h1>'''SDS-PAGE'''</h1> | <h1>'''SDS-PAGE'''</h1> | ||
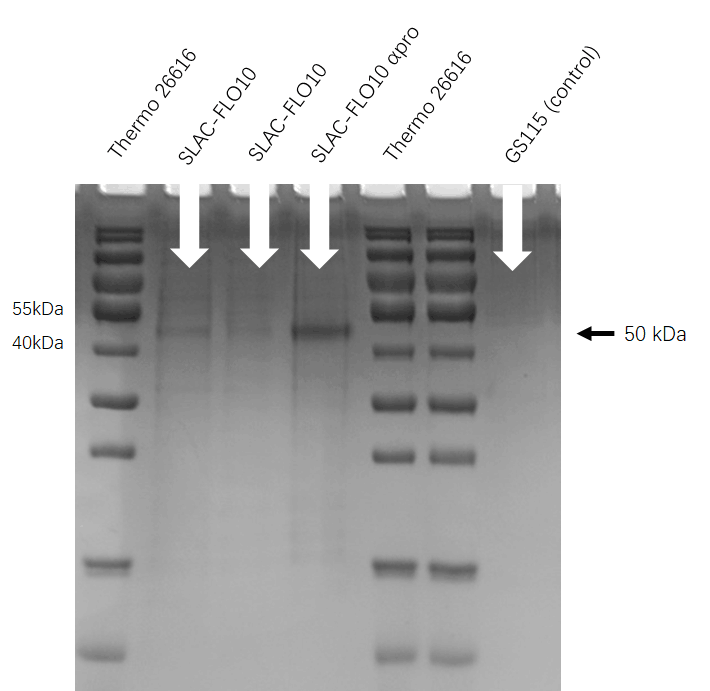
We run the SDS-PAGE to check whether PHO1 αpro help the enzyme transfer to the extracellular. As the figure shows, we get the protein type about xxx bp which means the SDS-PAGE is successful. | We run the SDS-PAGE to check whether PHO1 αpro help the enzyme transfer to the extracellular. As the figure shows, we get the protein type about xxx bp which means the SDS-PAGE is successful. | ||
| − | [[File: | + | [[File:T--HUST-China--2019-SLAC-SDS-PAGE.jpg |400px|thumb|center|Figure3:the SDS-page result shows that Pichia pastoris have successfully secrete SLAC protein into superfluous liquid.]] |
| + | |||
<h1>'''Enzyme Activity'''</h1> | <h1>'''Enzyme Activity'''</h1> | ||
We use ABTS to detect the enzyme activity. As the figure shows, the solution turns green, which confirm the enzyme activity. | We use ABTS to detect the enzyme activity. As the figure shows, the solution turns green, which confirm the enzyme activity. | ||
| − | [[File: | + | [[File:T--HUST-China--2019-SLAC enzyme activity.png|400px|thumb|center|Figure4:these four pictures shows the enzyme activity changes with the time. The four kinds of Pichia pastoris have the different curve, and the most activity strain is FLO10-apro, PHO1apro shows a more delayed increase on enzyme activity.]] |
| + | |||
<!-- --> | <!-- --> | ||
Revision as of 08:38, 20 October 2019
AOX1-Kozak-PHO1 pro-SLAC-His tag-AOX1 Terminator
PHO1 αpro is a combined signal peptide, which enhance the enzyme activity 3.5 times. SLAC can catalyze lignin.
Characterization
This is a four section for degrade and transfer lignin part.
DNA Gel Electrophoretic
After we link PHO1 αpro and SLAC successfully, we run the PCR with an intention to confirm the expression of SLAC. As the figure shows, we get the genetic stripe about 2019 bp which means the PCR is successful.
SDS-PAGE
We run the SDS-PAGE to check whether PHO1 αpro help the enzyme transfer to the extracellular. As the figure shows, we get the protein type about xxx bp which means the SDS-PAGE is successful.
Enzyme Activity
We use ABTS to detect the enzyme activity. As the figure shows, the solution turns green, which confirm the enzyme activity.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 937
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 1568