Difference between revisions of "Part:BBa K1334002"
| Line 26: | Line 26: | ||
Figure 3 The growth condition of E.coli strain DH5a after formaldehyde stimulation. | Figure 3 The growth condition of E.coli strain DH5a after formaldehyde stimulation. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
<br/> | <br/> | ||
| Line 37: | Line 32: | ||
==The effect of formaldehyde to the cells growth== | ==The effect of formaldehyde to the cells growth== | ||
<br/> | <br/> | ||
| − | Formaldehyde is toxic to the cells as we know, inhibiting | + | Formaldehyde is toxic to the cells as we know, inhibiting cells growth if it is added early to the culture medium. In order to know at which OD600 the formaldehyde should be added into the culture medium, we added the 0.5mM formaldehyde to cells at different original OD600, then we collected different group bacteria at certain time to measure cells’ DO600 absorbance (Figure 4). The result showed that the toxic effect of formaldehyde on the cells growth was decreased with the cells original OD600 increased. |
<br/> | <br/> | ||
==The sensitivity of the promoter to formaldehyde== | ==The sensitivity of the promoter to formaldehyde== | ||
<br/> | <br/> | ||
| − | Different concentration of formaldehyde has different ability to induce expression. In order to find the best concentration of formaldehyde to induce this promoter, we measured the sensitivity of the promoter to formaldehyde using BL21 as chassis. We cultured E.coli BL21 transformed with the plasmid (P-formaldehyde plus GFP)and added different concentration formaldehyde to different groups | + | Different concentration of formaldehyde has different ability to induce expression. In order to find the best concentration of formaldehyde to induce this promoter, we measured the sensitivity of the promoter to formaldehyde using BL21 as chassis. We cultured E.coli BL21 transformed with the plasmid (P-formaldehyde plus GFP)and added different concentration formaldehyde to different groups at OD600 = 1. Then we collected different group bacteria at certain time to measure the fluorescence and the absorbance (Figure 5). The result showed that 0.8 mM formaldehyde is the best concentration for inducing GFP expression. |
<br/> | <br/> | ||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_K1334002 SequenceAndFeatures</partinfo> | ||
Revision as of 03:01, 20 October 2019
HxlR+P-formaldehyde
A right promoter which can sense formaldehyde.There is a left promoter which can erpress the HxlR protein.The HxlR protein can enhance the function of the P-formaldehyde.
Usage and Biology
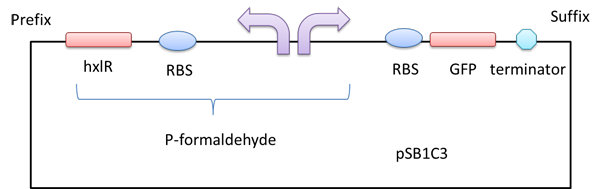
A gfp reporter gene was added downstream so that we can know whether the P-formaldehyde works or not in the presence of formaldehyde by detecting the relative fluorescence intensity.
Figure 1 A GFP reporter is added downstream of the formaldehyde promoter.
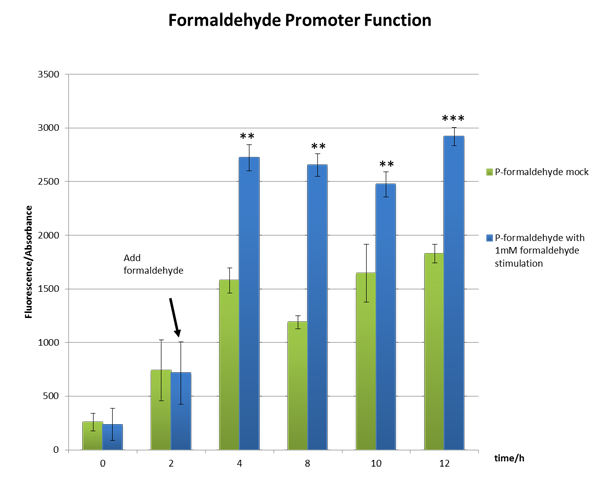
We incubated E.coli strain DH5a with the above plasmid (P-formaldehyde plus GFP)and stimulated the experimental group with 1mM formaldehyde when the OD600 value reached approximately one, which means E.coli comes into mid-log phase. And then we collected bacteria at certain time to measure the fluorescence and the absorbance.
From the Figure 2, we can see significant increase in relative fluorescence intensity after formaldehyde stimulation, which shows that our coloration system works well.
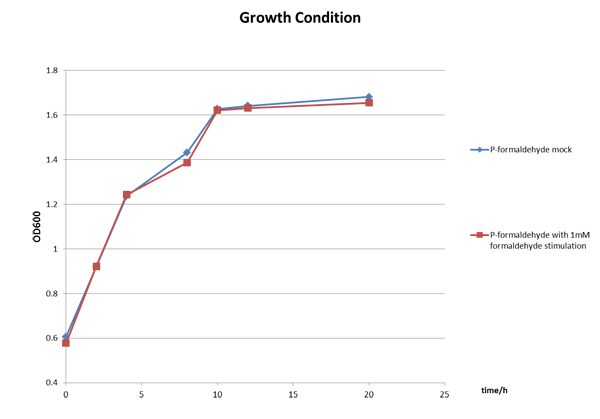
The Growth Condition curve in Figure 3 shows that the difference of the fluorescence/absorbance is not generated by the absorbance difference.
Figure 2 The function test of the formaldehyde promoter which shows our part BBa_K1334002 works well。
Figure 3 The growth condition of E.coli strain DH5a after formaldehyde stimulation.
Characterization from JNFLS 2019
The effect of formaldehyde to the cells growth
Formaldehyde is toxic to the cells as we know, inhibiting cells growth if it is added early to the culture medium. In order to know at which OD600 the formaldehyde should be added into the culture medium, we added the 0.5mM formaldehyde to cells at different original OD600, then we collected different group bacteria at certain time to measure cells’ DO600 absorbance (Figure 4). The result showed that the toxic effect of formaldehyde on the cells growth was decreased with the cells original OD600 increased.
The sensitivity of the promoter to formaldehyde
Different concentration of formaldehyde has different ability to induce expression. In order to find the best concentration of formaldehyde to induce this promoter, we measured the sensitivity of the promoter to formaldehyde using BL21 as chassis. We cultured E.coli BL21 transformed with the plasmid (P-formaldehyde plus GFP)and added different concentration formaldehyde to different groups at OD600 = 1. Then we collected different group bacteria at certain time to measure the fluorescence and the absorbance (Figure 5). The result showed that 0.8 mM formaldehyde is the best concentration for inducing GFP expression.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]