Difference between revisions of "Part:BBa B0034"
| Line 14: | Line 14: | ||
<p>Biobrick RBSs [https://parts.igem.org/Part:BBa_K1956022 B0030], [https://parts.igem.org/Part:BBa_K1956023 B0031], [https://parts.igem.org/Part:BBa_K1956014 B0032], [https://parts.igem.org/Part:BBa_K1956015 B0034] were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.</p> | <p>Biobrick RBSs [https://parts.igem.org/Part:BBa_K1956022 B0030], [https://parts.igem.org/Part:BBa_K1956023 B0031], [https://parts.igem.org/Part:BBa_K1956014 B0032], [https://parts.igem.org/Part:BBa_K1956015 B0034] were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.</p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
Note: The Elowitz RBS is the definition of efficiency 1.0. | Note: The Elowitz RBS is the definition of efficiency 1.0. | ||
| Line 73: | Line 67: | ||
|0.398 | |0.398 | ||
|} | |} | ||
| + | |||
| + | |||
| + | <!-- --> | ||
| + | <span class='h3bb'>Sequence and Features</span> | ||
| + | <partinfo>BBa_B0034 SequenceAndFeatures</partinfo> | ||
| + | |||
| + | |||
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_B0034 parameters</partinfo> | ||
Revision as of 15:01, 19 October 2019
RBS (Elowitz 1999) -- defines RBS efficiency
RBS based on Elowitz repressilator.
Usage and Biology
IIT Madras 2016's Characterization
Modelling
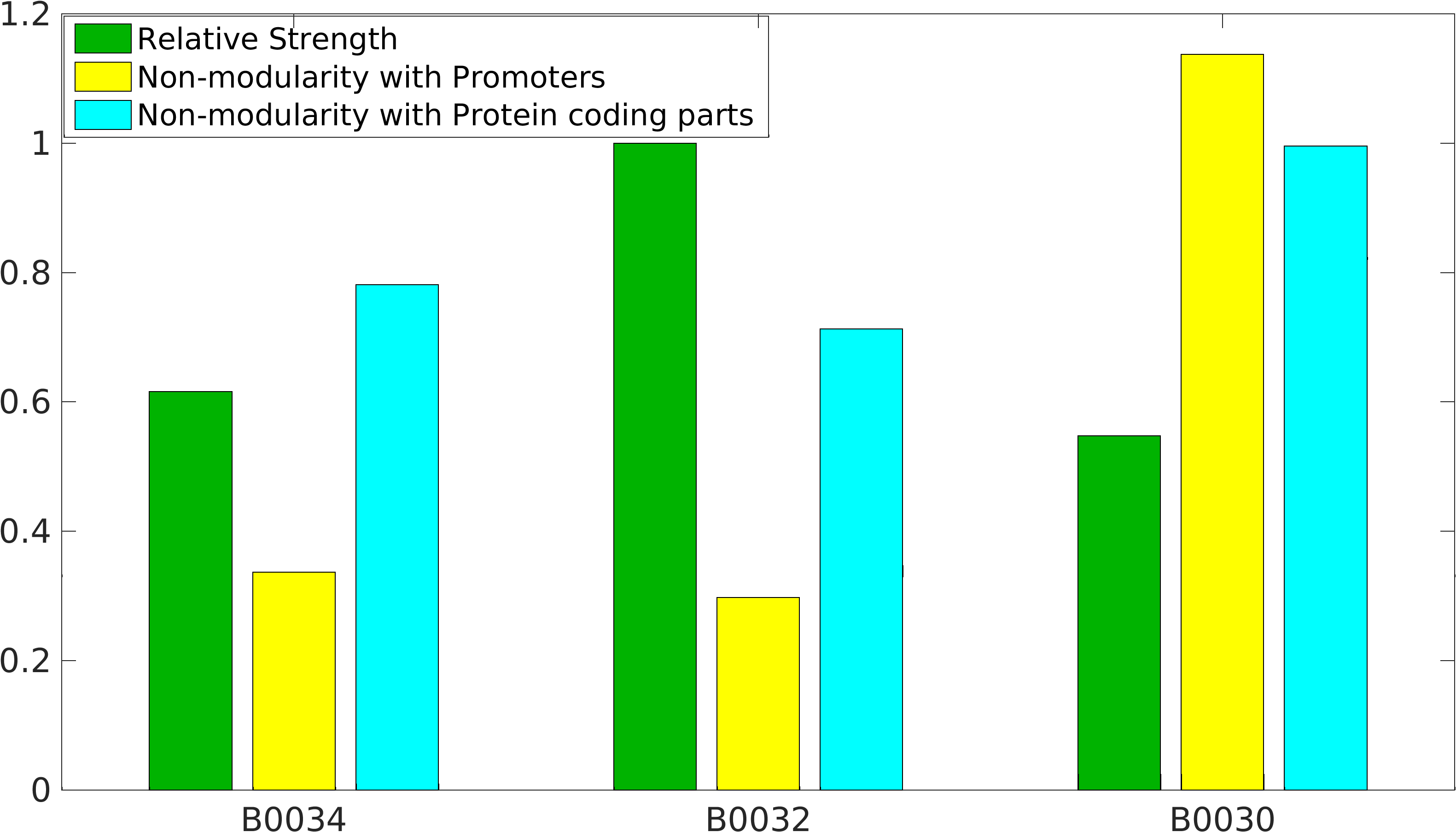
Global non-modularity towards promoters & protein coding parts and relative strength was estimated for RBSs B0030, B0032, B0034 in our [http://2016.igem.org/Team:IIT-Madras/Model#Modularity_of_RBS_parts modelling]
Experimentation
Biobrick RBSs B0030, B0031, B0032, B0034 were used in our 'Noise in Device' experiment to understand the role of RBS parts in giving rise to noise.
Note: The Elowitz RBS is the definition of efficiency 1.0.
Team Warsaw 2010's measurement
definition of 100% strength in our measurement.Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have done the characterization of this RBS.
Documentation:
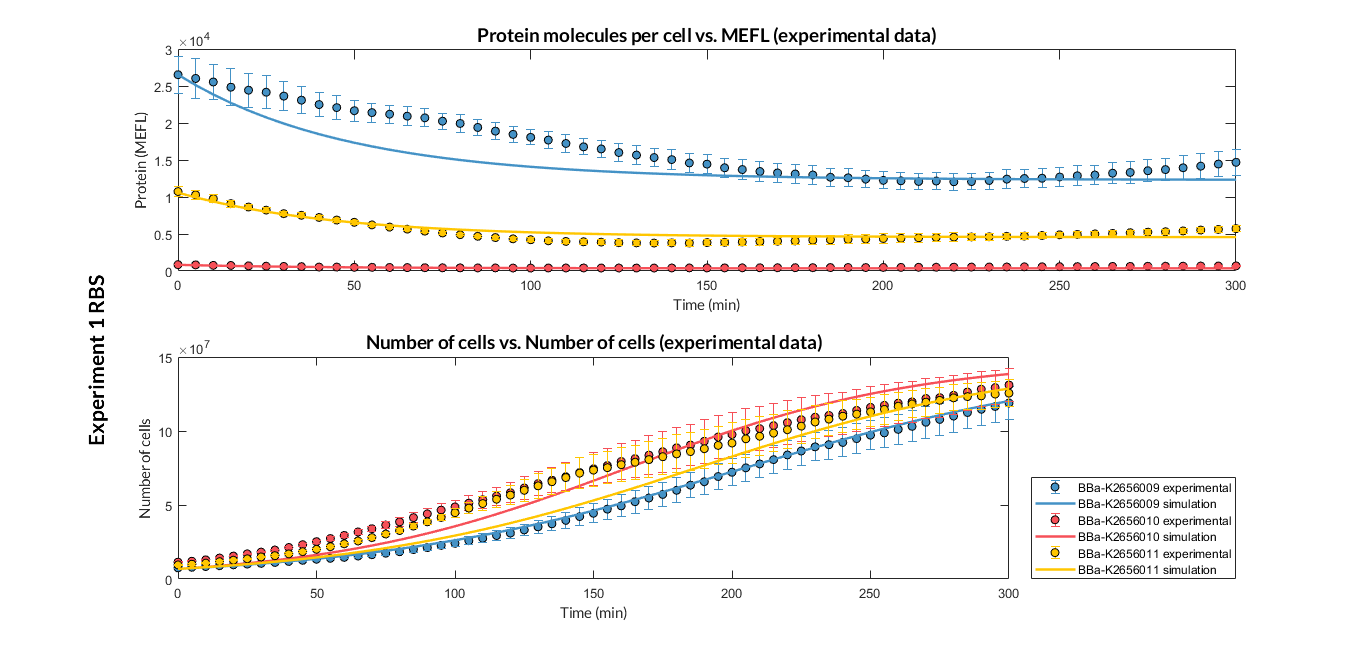
BBa_K2656011 is the RBS B0034 standardized into the Golden Braid assembly method. It also includes the BioBrick equivalent scar in the 3' extreme, so the insertion of this supplementary bases ensure correct spacing for the CDS expression when assembled into a TU.
Characterization of the this part was performed with the transcriptional unit BBa_K2656101 , which was used in a comparative RBS expression experiment with composite parts BBa_K2656101 and BBa_K2656102. They all were assembled in a Golden Braid alpha1 plasmid using the same promoter, CDS and terminator.
By using this [http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model]and rationale choose its [http://2018.igem.org/Team:Valencia_UPV/Modeling#optimization optimization values] based on each RBS tested.
| Table 1. Optimized parameters from experimental data (BBa_K2656009 RBS). | |||
| Parameter | Value | ||
| Translation rate p | p = 0.082 min-1 | ||
| Dilution rate μ | μ = 0.01631 min-1 | ||
We have also calculated the relative force between the different RBS, taking BBa_K2656009 strong RBS as a reference. Likewise, a ratio between p parameters of the different RBS parts and p parameter of the reference RBS has been calculated.
| Table 2. BBa_K2656008 (GB BBa_B0032 RBS) relative strength and p ratio. | |||
| Parameter | Value | ||
| Relative strength | 0.371 | ||
| p parameter ratio (pRBS/pref) | 0.398 | ||
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Functional Parameters
| biology | -NA- |
| efficiency | 1 |