Difference between revisions of "Part:BBa K165000"
| (10 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
| − | + | The is a repressible promoter for use in <i>Saccharomyces cerevisiae</i>. It is repressed by the presence of methionine. When unrepressed, is has a moderate level of activity. | |
<!-- Add more about the biology of this part here | <!-- Add more about the biology of this part here | ||
| Line 18: | Line 18: | ||
<!-- --> | <!-- --> | ||
INTRODUCTION | INTRODUCTION | ||
| − | <p>In order to determine the strength of pMET25 promoter, we have conducted the following experiment. First of all, we have constructed | + | <p>pMET25 is inducible(silent when methionine is present in the growth medium and is activated in methionine drop-out medium). In order to determine the strength of pMET25 promoter, we have conducted the following experiment. First of all, we have constructed a plasmid where EGFP is expressed under MET25 promoter. The empty plasmid was used as a negative control. |
| − | </p><p> | + | </p><p>Methodology |
| − | + | ||
| − | + | All common methods such as PCR, agarose gel electrophoresis, miniprep, bacterial and yeast transformation, and some others are described in the Methodology section in Tartu_TUIT2019 wiki page. | |
| − | + | ||
| − | + | Plasmid construction | |
| − | + | Target promoters were PCR amplified from yeast genomic DNA with primers containing SacI (forward primer) and NotI (reverse primer) restriction sites at their 5’-ends. PCR products were separated on the agarose gel, purified and cut with SacI/NotI enzymes. As a backbone, we used pRS306 plasmid containing pTDH3-GFP-tCYC1 cassette. The vector was digested with SacI/NotI restriction enzymes, which cut out the pTDH3 promoter region, and was ligated with our target promoter. After bacterial transformation and miniprep, sequence-verified plasmids were used for yeast transformation. | |
| − | + | ||
| − | + | Yeast strain construction | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | Constructed vectors with GFP under target promoters were restricted and used for yeast transformation.<i>S. cerevisiae</i> DOM90 (MATa <i>{leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 bar1::hisG} [phi+])</i> strain was transformed. All the yeast strains generated and used for promoter characterization are listed in Table 1. | |
| + | |||
| + | Table 1. Yeast strains used for promoter characterization experiments | ||
| − | |||
{| class="wikitable" | {| class="wikitable" | ||
| − | + | |Strain name | |
| − | |Strain name | + | |Genotype* |
| − | |Genotype | + | |Description |
| − | | | + | |
|- | |- | ||
| − | | | + | |DOM0090 |
| − | | | + | |<i>ura3</i>-1::pRS306-URA3 |
| − | + | |Strain with empty vector; was used as a negative control for GFP fluorescence | |
| − | | | + | |
|- | |- | ||
| − | | | + | |TTI_1 |
| − | | | + | |<i>ura3</i>-1::pRS306-pTDH3-EGFP-tCYC1-URA3 |
| − | + | |Strain with EGFP under TDH3 promoter; was used as a positive control for GFP fluorescence | |
| − | + | ||
|- | |- | ||
| − | | | + | |TTI_2 |
| − | | | + | |<i>ura3</i>-1::pRS306 pMET25-EGFP-tCYC1-URA3 |
| − | + | |Strain with GFP under pMET25 promoter | |
| − | + | ||
|} | |} | ||
| − | + | *DOM90 strain (MATa <i>{leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 bar1::hisG} [phi+])</i> was used for transformation. In the Genotype column, only the differences between strains are provided. | |
| + | Following transformation, separate yeast colonies were screened for the presence of the insert under the fluorescent microscope. Colonies displaying GFP fluorescence were selected for further fluorescence measurement experiment. | ||
| − | + | Fluorescence measurement | |
| − | + | Prior to measurements, yeast strains were pregrown in liquid media overnight at 30°C and 200 RPM in the shaker incubator. The next day, optical density (OD600) of yeast cultures was measured. Proper culture volumes were first centrifuged, washed with distilled water and resuspended in either inductive or non-inductive media to get final culture OD600 from 0.8 to 1.1. Before the fluorescence measurements, cell cultures were grown for 3 hours at 30°C and 200 RPM in the shaker incubator. | |
| + | All the media used for different strains are listed in Table 2. | ||
| + | |||
| + | Table 2. Media used for cell growth | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | | | + | |Strain |
| − | | | + | |Medium |
| − | | | + | |Carbon source |
| − | + | ||
| − | + | ||
|- | |- | ||
| − | | | + | |<i>ura3</i>-1::pRS306-URA3 |
| − | + | |CSM | |
| − | + | |2% glucose | |
| − | | | + | |
| − | | | + | |
|- | |- | ||
| − | | | + | |<i>ura3</i>-1::pRS306-pTDH3-EGFP-tCYC1-URA3 |
| − | + | |CSM | |
| − | + | |2% glucose | |
| − | | | + | |
| − | | | + | |
|- | |- | ||
| − | | | + | |<i>ura3</i>-1::pRS306 pMET25-EGFP-tCYC1-URA3 |
| − | + | |CSM (non-inductive conditions) | |
| − | + | CSM methionine drop-out(inductive conditions) | |
| − | + | |2% glucose | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | | | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
| − | | | + | |
| − | + | ||
| − | + | ||
| − | + | ||
| − | + | ||
|} | |} | ||
| − | + | After 3 hours of incubation, 200 µL of each strain culture was transferred into a 96-well plate (clear flat bottom). Two biological replicates (two different positive colonies after transformation) of each genotype were taken in four technical replicates for fluorescence measurements. | |
| + | For the fluorescence measurement, we have used BioTek Synergy MX microplate reader with the following setup: excitation 485 nm, emission 528 nm, bandwidth 20, gain 80. Alone with fluorescence, absorbance at 600 nm was measured (with disabled pathlength correction). CSM medium was used as a blank for OD600 measurements. Fluorescein was used as a reference (according to the protocol of 2018 iGEM InterLab Study) to quantify GFP fluorescence intensity. However, in our case, the standard curve for fluorescein concentrations in the range from 0.0012 to 0.313 µM was generated, in contrast to the InterLab Study, where 10 µM of fluorescein was the highest concentration. Since we used a higher gain parameter in our experiment, fluorescein concentrations above 0.313 µM showed intensities that were above the detection limit of our device. | ||
| + | Results | ||
| + | EGFP was cloned under target promoter and constructs were transformed into yeast cells. The intensity of GFP fluorescence was taken as a measure of promoter strength. | ||
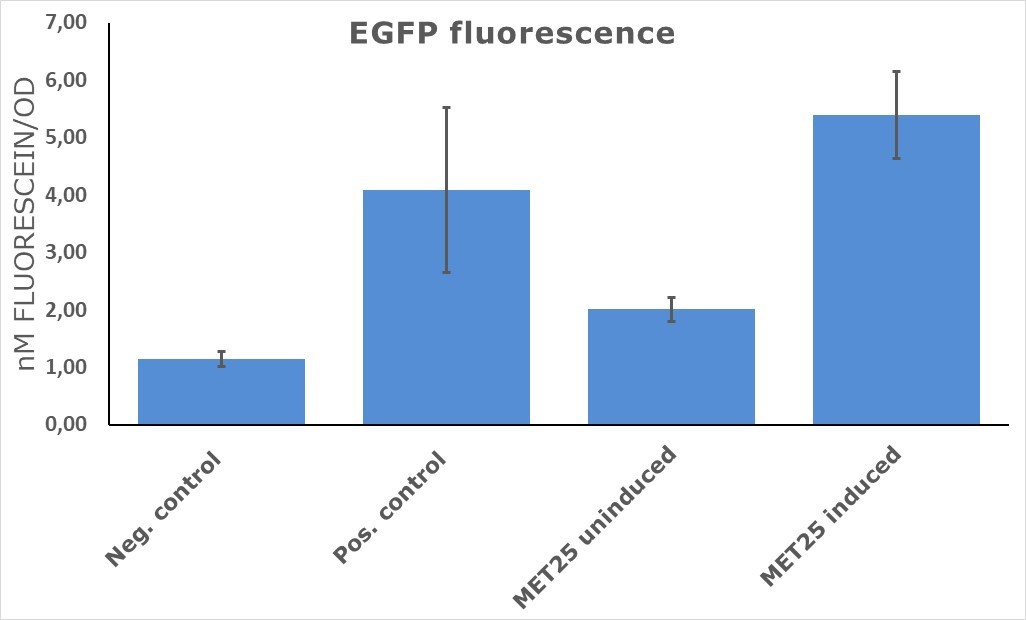
| + | As can be seen in figure 1, EGFP under induced pMet25 showed 40% higher fluorescence in comparison to a positive control (pTDH3-EGFP). Surprisingly, EGFP fluorescence was detected in the strains with the inducible promoter under non-inductive conditions (presence of methionine). | ||
| − | + | Figure 1. | |
| − | + | [[File:uninMet25.jpg|500px]] | |
CONCLUSIONS | CONCLUSIONS | ||
| − | + | ||
| + | The results of EGFP fluorescence measurements suggest that characterized yeast promoter initiate transcription at the rate, which was higher in our experiment than one observed for pTDH3 (positive control). Surprisingly, pMET25 promoter showed some leakage effect under non-inductive conditions that should be taken into consideration while choosing a promoter for the experiment. | ||
Latest revision as of 11:35, 19 October 2019
MET 25 Promoter
The is a repressible promoter for use in Saccharomyces cerevisiae. It is repressed by the presence of methionine. When unrepressed, is has a moderate level of activity.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
INTRODUCTION
pMET25 is inducible(silent when methionine is present in the growth medium and is activated in methionine drop-out medium). In order to determine the strength of pMET25 promoter, we have conducted the following experiment. First of all, we have constructed a plasmid where EGFP is expressed under MET25 promoter. The empty plasmid was used as a negative control.
Methodology
All common methods such as PCR, agarose gel electrophoresis, miniprep, bacterial and yeast transformation, and some others are described in the Methodology section in Tartu_TUIT2019 wiki page.
Plasmid construction Target promoters were PCR amplified from yeast genomic DNA with primers containing SacI (forward primer) and NotI (reverse primer) restriction sites at their 5’-ends. PCR products were separated on the agarose gel, purified and cut with SacI/NotI enzymes. As a backbone, we used pRS306 plasmid containing pTDH3-GFP-tCYC1 cassette. The vector was digested with SacI/NotI restriction enzymes, which cut out the pTDH3 promoter region, and was ligated with our target promoter. After bacterial transformation and miniprep, sequence-verified plasmids were used for yeast transformation.
Yeast strain construction
Constructed vectors with GFP under target promoters were restricted and used for yeast transformation.S. cerevisiae DOM90 (MATa {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 bar1::hisG} [phi+]) strain was transformed. All the yeast strains generated and used for promoter characterization are listed in Table 1.
Table 1. Yeast strains used for promoter characterization experiments
| Strain name | Genotype* | Description |
| DOM0090 | ura3-1::pRS306-URA3 | Strain with empty vector; was used as a negative control for GFP fluorescence |
| TTI_1 | ura3-1::pRS306-pTDH3-EGFP-tCYC1-URA3 | Strain with EGFP under TDH3 promoter; was used as a positive control for GFP fluorescence |
| TTI_2 | ura3-1::pRS306 pMET25-EGFP-tCYC1-URA3 | Strain with GFP under pMET25 promoter |
- DOM90 strain (MATa {leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 bar1::hisG} [phi+]) was used for transformation. In the Genotype column, only the differences between strains are provided.
Following transformation, separate yeast colonies were screened for the presence of the insert under the fluorescent microscope. Colonies displaying GFP fluorescence were selected for further fluorescence measurement experiment.
Fluorescence measurement
Prior to measurements, yeast strains were pregrown in liquid media overnight at 30°C and 200 RPM in the shaker incubator. The next day, optical density (OD600) of yeast cultures was measured. Proper culture volumes were first centrifuged, washed with distilled water and resuspended in either inductive or non-inductive media to get final culture OD600 from 0.8 to 1.1. Before the fluorescence measurements, cell cultures were grown for 3 hours at 30°C and 200 RPM in the shaker incubator. All the media used for different strains are listed in Table 2.
Table 2. Media used for cell growth
| Strain | Medium | Carbon source |
| ura3-1::pRS306-URA3 | CSM | 2% glucose |
| ura3-1::pRS306-pTDH3-EGFP-tCYC1-URA3 | CSM | 2% glucose |
| ura3-1::pRS306 pMET25-EGFP-tCYC1-URA3 | CSM (non-inductive conditions)
CSM methionine drop-out(inductive conditions) |
2% glucose |
After 3 hours of incubation, 200 µL of each strain culture was transferred into a 96-well plate (clear flat bottom). Two biological replicates (two different positive colonies after transformation) of each genotype were taken in four technical replicates for fluorescence measurements. For the fluorescence measurement, we have used BioTek Synergy MX microplate reader with the following setup: excitation 485 nm, emission 528 nm, bandwidth 20, gain 80. Alone with fluorescence, absorbance at 600 nm was measured (with disabled pathlength correction). CSM medium was used as a blank for OD600 measurements. Fluorescein was used as a reference (according to the protocol of 2018 iGEM InterLab Study) to quantify GFP fluorescence intensity. However, in our case, the standard curve for fluorescein concentrations in the range from 0.0012 to 0.313 µM was generated, in contrast to the InterLab Study, where 10 µM of fluorescein was the highest concentration. Since we used a higher gain parameter in our experiment, fluorescein concentrations above 0.313 µM showed intensities that were above the detection limit of our device.
Results
EGFP was cloned under target promoter and constructs were transformed into yeast cells. The intensity of GFP fluorescence was taken as a measure of promoter strength. As can be seen in figure 1, EGFP under induced pMet25 showed 40% higher fluorescence in comparison to a positive control (pTDH3-EGFP). Surprisingly, EGFP fluorescence was detected in the strains with the inducible promoter under non-inductive conditions (presence of methionine).
CONCLUSIONS