Difference between revisions of "Part:BBa K2912016"
m (→SZU-China 2019 iGEM team) |
m (→SZU-China 2019 iGEM team) |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 19: | Line 19: | ||
<div> | <div> | ||
<center><html><img src='https://2019.igem.org/wiki/images/3/3e/T--SZU-CHINA--tofig4.jpg' style="width:90%;margin:0 auto"> | <center><html><img src='https://2019.igem.org/wiki/images/3/3e/T--SZU-CHINA--tofig4.jpg' style="width:90%;margin:0 auto"> | ||
| − | <center> Fig.1 Schematic representation of the direct detection of miRNAs </center></html></center> | + | <center> <span style="font-weight:bold">Fig.1 Schematic representation of the direct detection of miRNAs</span> </center></html></center> |
</div> | </div> | ||
| − | This year, we first introduced this strategy to test the function of toxin Tse2 and finally verified that Tse2 could work, and this testing method was useful and reliable. For more information, please visit [https://parts.igem.org/Part:BBa_K314200 BBa_K314200 | + | This year, we first introduced this strategy to test the function of toxin Tse2 and finally verified that Tse2 could work, and this testing method was useful and reliable. For more information, please visit [https://parts.igem.org/Part:BBa_K314200 BBa_K314200.] |
| − | The G-quadruplexes DNA in this page was designed to detect the siRNA inside ''M. micrantha'' leaves to test the efficiency of our RNAi-based heribicide for ''M. micrantha''. | + | The G-quadruplexes DNA in this page was designed to detect the siRNA inside ''M. micrantha'' leaves to test the efficiency of our RNAi-based heribicide for ''M. micrantha'' (Fig.2). |
| + | |||
| + | <div> | ||
| + | <center><html><img src='https://2019.igem.org/wiki/images/5/5c/T--SZU-CHINA--siRNA_detect_comparison.png' style="width:90%;margin:0 auto"> | ||
| + | <center> <span style="font-weight:bold">Fig.2 The siRNA detection results vs the RT-qPCR results</span> </center></html></center> | ||
| + | </div> | ||
| + | |||
| + | As shown in Fig.2, we can see that target genes were significantly silenced after treated for 2 days and recovered after about 4 days according to the RT-qPCR results, while the siRNA in vivo increased dramatically after treated for 1 days and then decreased gradually according to the new method results, from which we can conclude that with the increase of siRNA in vivo, the gene expression was down-regulated as expected and the decrease of siRNA led to the recovery of target gene expression. Hence, compared with the results of RT-qPCR, which is a traditional way to test the gene expression of target gene, the siRNA detection results are probably reliable and can reflect the change of siRNA in vivo to some extent. | ||
| + | |||
| + | This G-rich DNA duplex was designed to test the change of siRNA named 29128. For more details, please see [https://parts.igem.org/Part:BBa_K2912011 BBa_K2912011.] | ||
===References=== | ===References=== | ||
Latest revision as of 00:19, 19 October 2019
G-quadruplexes used to detect miRNA
MicroRNAs (miRNAs) have been considered to be potent biomarkers for early disease diagnosis and for cancer therapy. The rapid and selective detection of miRNAs without reverse transcription and labelling is highly desired. Herein, we reported a simple and label-free miRNA detection method.The interaction of the G-quadruplex DNA structure with Thioflavin T (ThT) led to a label-free signal output. Under the optimised conditions, this method allowed for simple, rapid, and sequence-specific detection of miRNA. SZU-China this year used G-quadruplexes to detect siRNA introduced into the leaves of M. micrantha quantatively.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
SZU-China 2019 iGEM team
SZU-China 2019 iGEM team this year aimed to develop an RNAi-based herbicide for M. micrantha. We constructed a plasmid and transformed it into E. coli ht115 (DE3), which then could transcribe hairpin siRNA. We found a G-quadruplex DNA-based, label-free and ultrasensitive strategy to detect the siRNA.
A cDNA strand, which is completely complementary to the target miRNA and partly complementary pairing with G-rich DNA, was designed first. Then this cDNA can be competed off from the cDNA/G-rich DNA duplex to form a cDNA/RNA heteroduplex and release the G-rich oligonucleotides when the target-siRNA was introduced. Recent research progress has demonstrated that G-quadruplex DNA, a specific type of G-rich nucleic acid sequence, can be remarkably recognized by thioflavin T (ThT) with high selectivity, unlike the triplex, duplex or single-stranded forms of DNA. The fluorescence intensity of ThT exhibits a considerable increase upon binding to G-quadruplex DNA, which can be utilized as a signal reporter. The conformation of released G-rich oligonucleotides would change into G-quadruplex DNA with the presence of 2.0 mM K+. Then, the ThT remarkably recognizes and selectively binds to the G-quadruplex DNA, resulting in a significant enhancement in the fluorescence signal (Fig.1).

This year, we first introduced this strategy to test the function of toxin Tse2 and finally verified that Tse2 could work, and this testing method was useful and reliable. For more information, please visit BBa_K314200.
The G-quadruplexes DNA in this page was designed to detect the siRNA inside M. micrantha leaves to test the efficiency of our RNAi-based heribicide for M. micrantha (Fig.2).
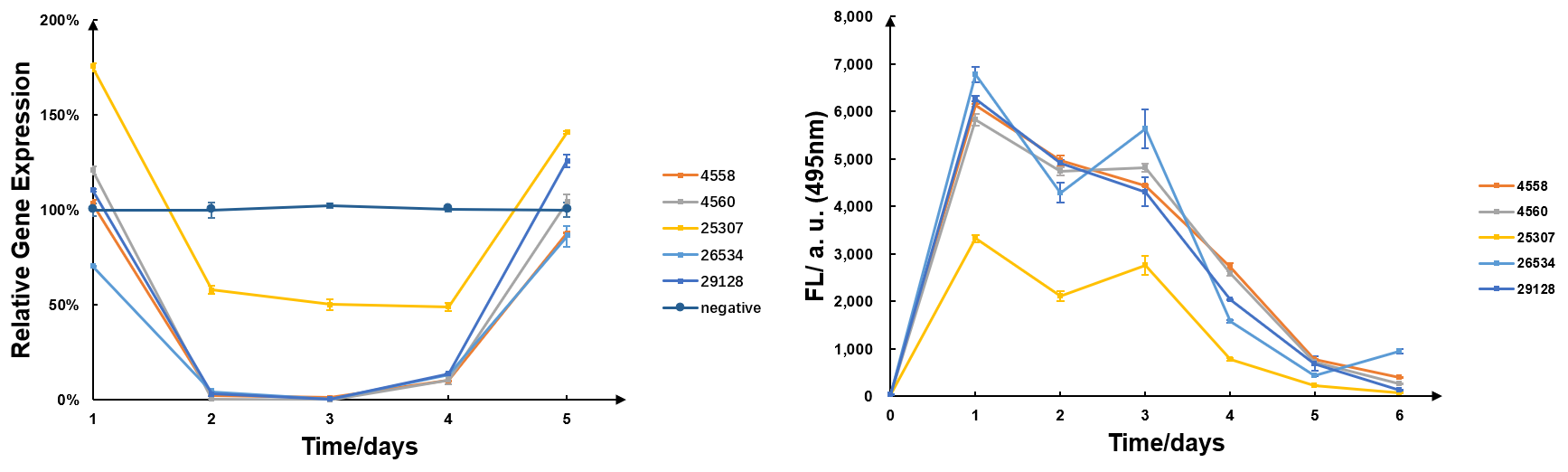
As shown in Fig.2, we can see that target genes were significantly silenced after treated for 2 days and recovered after about 4 days according to the RT-qPCR results, while the siRNA in vivo increased dramatically after treated for 1 days and then decreased gradually according to the new method results, from which we can conclude that with the increase of siRNA in vivo, the gene expression was down-regulated as expected and the decrease of siRNA led to the recovery of target gene expression. Hence, compared with the results of RT-qPCR, which is a traditional way to test the gene expression of target gene, the siRNA detection results are probably reliable and can reflect the change of siRNA in vivo to some extent.
This G-rich DNA duplex was designed to test the change of siRNA named 29128. For more details, please see BBa_K2912011.
References
[1] Liu S, Peng P, Wang H, et al. Thioflavin T binds dimeric parallel-stranded GA-containing non-G-quadruplex DNAs: a general approach to lighting up double-stranded scaffolds.[J]. Nucleic Acids Research, 2017.
[2] Yan L, Yan Y, Pei L, et al. A G-quadruplex DNA-based, Label-Free and Ultrasensitive Strategy for microRNA Detection[J]. Scientific Reports, 2014, 4:7400.
