Difference between revisions of "Part:BBa K592101"
| Line 145: | Line 145: | ||
</html> | </html> | ||
| + | <html> | ||
<br> | <br> | ||
<br> | <br> | ||
| Line 155: | Line 156: | ||
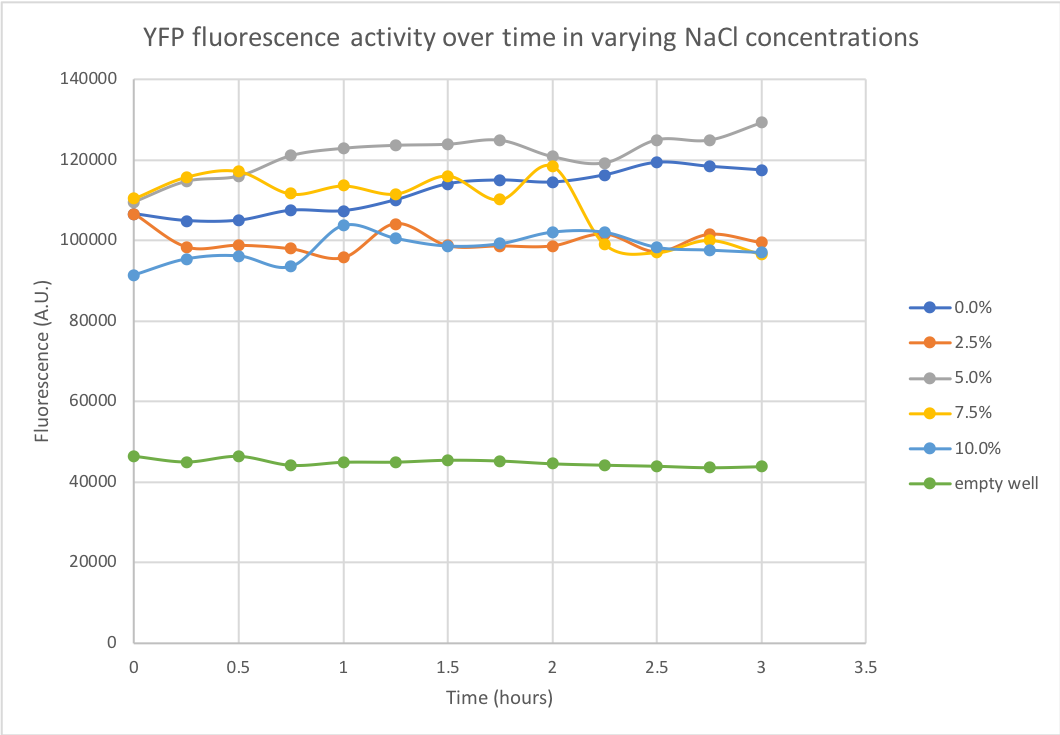
Documentation: We performed a kinetic measurement of YFP degradation in different salt concentrations over 3 hours using a spectrofluorometer, with measurements being taken every 15 minutes. Once the experiment was carried out, the results were plotted in Figure 1. in which we can observe that the protein remains quite stable in salt concentrations as high as 10% over a 3 hour period. | Documentation: We performed a kinetic measurement of YFP degradation in different salt concentrations over 3 hours using a spectrofluorometer, with measurements being taken every 15 minutes. Once the experiment was carried out, the results were plotted in Figure 1. in which we can observe that the protein remains quite stable in salt concentrations as high as 10% over a 3 hour period. | ||
<br> | <br> | ||
| − | + | </html> | |
[[File:T--KU_LEUVEN--YFP_graph_parts.png|900px|thumb|none|alt=YFP_graph.|Figure 3. YFP activity in the presence of NaCl over time.]] | [[File:T--KU_LEUVEN--YFP_graph_parts.png|900px|thumb|none|alt=YFP_graph.|Figure 3. YFP activity in the presence of NaCl over time.]] | ||
Revision as of 20:27, 16 October 2019
Yellow Fluorescent Protein (YFP)
Yellow Fluorescent Protein derived from Aequorea victoria GFP. This sequence is cloned from the pZE12-YFP plasmid used by Elowitz (see reference). The original gene was made by the The Yeast Resource Center (YRC) based at the University of Washington in Seattle, Washington.
iGEM12_Uppsala_University: If you are looking for a bright yellow fluorescent protein, the improved gene SYFP2 BBa_K864100 is a better choice than this part.
Usage and Biology
This part is useful as a reporter.
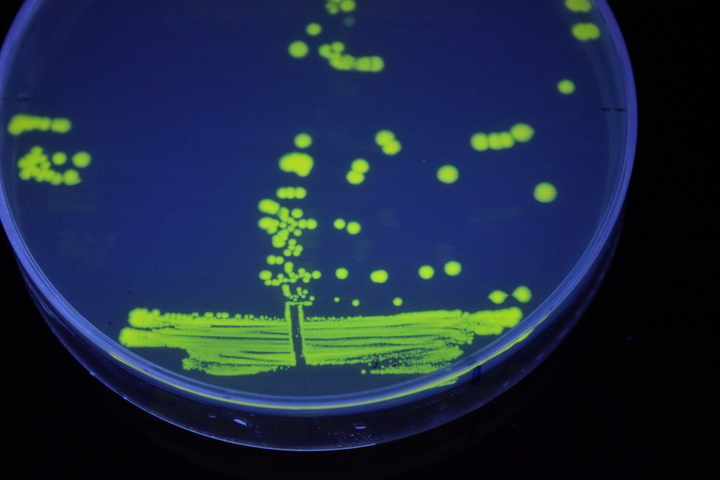
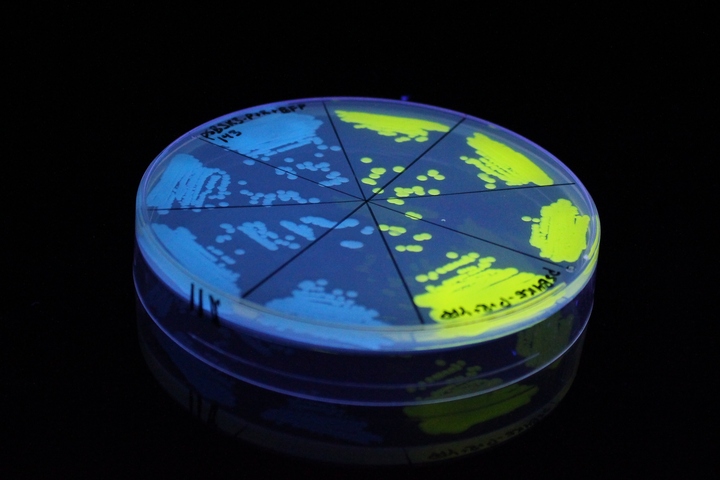
The images above show E coli constitutively expressing YFP BBa_K592101 (yellow) and mTagBFP BBa_K592100 (blue) illuminated on a UV table.
[http://2017.igem.org/Team:AFCM-Egypt# Egypt-AFCM Team] tried to improve YFP gel characterization and function at BBa_K592101 regarding its expression by the lac promoter (BBa_K2217017) as a weak constitutive promoter in one composite part to help enhancing YFP expression to characterize non-coding RNA regulatory activity. Part characterization and usage can be found at BBa_K2217023_Experience.
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez
Summary:We have adapted the part to be able to assemble transcriptional units with the Golden Gate method and we have added the degradation tag ssRA LVA. After that, we have characterized the change in the protein degradation due to this tag.
Documentation:
First, we adapted the CDS BBa_K592101 to be used to assemble composite parts using the Golden Gate method, creating BBa_K2656021 and we added the LVA degradation tag, creating BBa_K2656020, our improved part. Next, we performed an experiment to obtain the excitation and emission spectra. To do this, we created the transcriptional unit BBa_K2656112 and we used the parameters of the Table 1:
BBa_K2656004: the J23106 promoter in its GoldenBraid compatible version from our Part Collection
BBa_K2656009: the B0030 ribosome biding site in its GoldenBraid compatible version from our Part Collection
BBa_K2656021: The original part BBa_K592101 compatible with the GoldenBraid assembly method
BBa_K2656026: the B0015 transcriptional terminator in its GoldenBraid compatible version from our Part Collection
Table 1. Parameters used to obtain the spectra
| Parameter | Value |
| Number of samples | 6 |
| Excitation Wavelength measurement range (nm) | [450-550] |
| Emission wavelenght (nm) | 580 |
| Emission Wavelength measurement range (nm) | [500-580] |
| Excitation wavelenght (nm) | 470 |
| Gain (G) | 50 |
To test the effect of the degradation tag, we designed an experiment with which we measured the increase in protein degradation due to this tag. To perform this experiment, we assembled two composite parts with the same promoter, RBS and terminator:
BBa_K2656020: The original part with the added LVA ssRA degradation tag compatible with the GoldenBraid assembly method
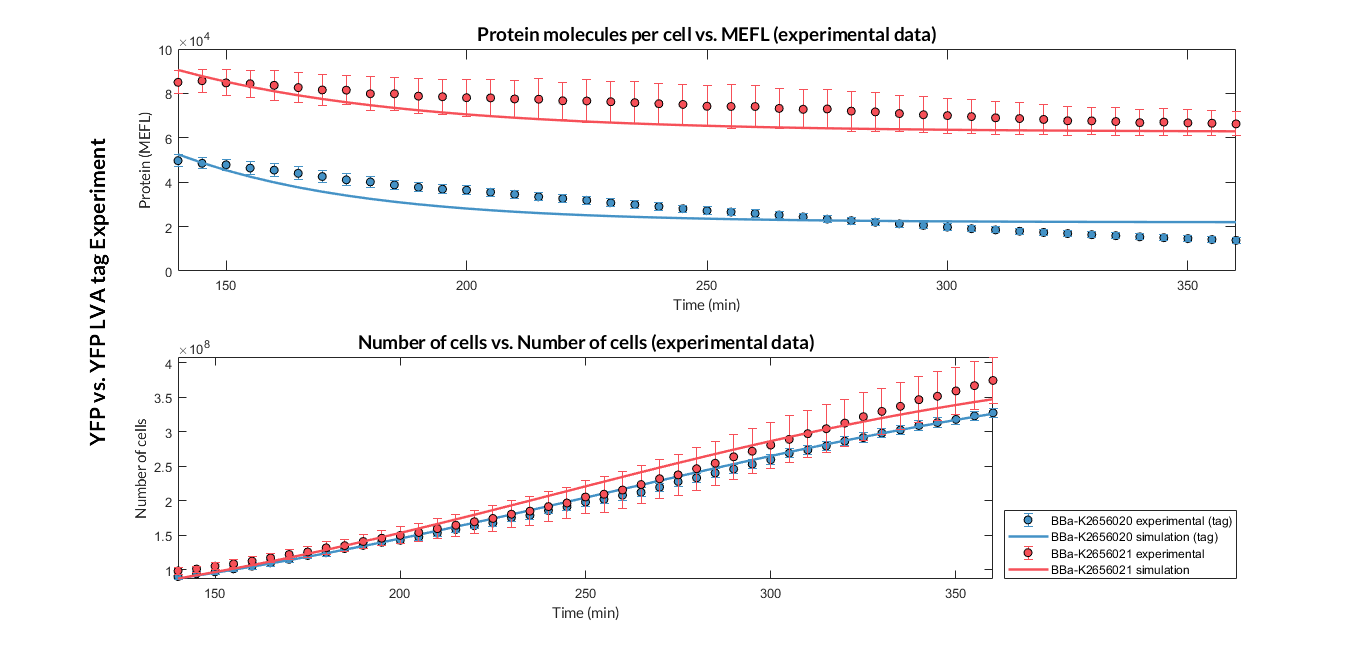
Once the experiment was carried out, the results were plotted and Figure 2 was obtained, in which we can observe that the growth of the bacteria with both constructions was very similar, while the fluorescence had a clear variation.
These data were optimized with our model and the parameters from Table 2 were obtained. With these parameters it is possible to obtain that the degradation of the protein with the tag is around twice as much as the one without the tag.
Table 2. Optimized values of translation rate, degradation rate and dilution rate from experimental data
Optimized parameters |
Values |
|---|---|
Translation rate p |
|
PoI degradation rate dp |
|
Dilution rate μ |
|
Group: KU_LEUVEN iGEM 2019
Authors: Zarina Nauryzgaliyeva, Cheska Cadacio
Summary: Our team has decided to investigate the properties of YFP (yellow fluorescent protein) in varying salt concentrations. The team has investigated YFP fluorescent activity at NaCl concentration of 0, 2.5, 5, 7.5and 10%, where 3% is the physiological salt concentration of seawater. The performed experiment and obtained data were beneficial for our project, where we envisioned to use marine cyanobacteria as a platform for enzyme production, therefore the stability of potentially secreted proteins is interesting to know. From literature we found GFP stability in different temperatures and in the presence of sodium chloride, therefore we wanted to obtain similar information about YFP(1).
Documentation: We performed a kinetic measurement of YFP degradation in different salt concentrations over 3 hours using a spectrofluorometer, with measurements being taken every 15 minutes. Once the experiment was carried out, the results were plotted in Figure 1. in which we can observe that the protein remains quite stable in salt concentrations as high as 10% over a 3 hour period.
References
[http://www.ncbi.nlm.nih.gov/pubmed/12183631] Elowitz, M. B., A. J. Levine, et al. (2002). "Stochastic gene expression in a single cell." Science 297(5584): 1183-6.
[1]Ishii M, Kunimura S, Tallon Jeng H, Vessoni Penna TC, Cholewa O. Evaluation of the pH- and Thermal Stability of the Recombinant Green Fluorescent Protein (GFP) in the Presence of Sodium Chloride. Appl Biochem Biotechnol. 2007;136:555–71.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 644