Difference between revisions of "Part:BBa K206000"
m (→Usage and Biology) |
|||
| (10 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| − | |||
| + | {| | ||
| + | |<div style="align: center; valign: center; font-family: Arial; font-size: 12pt"> | ||
| + | {| | ||
| + | |- | ||
| + | |width="60px"|''Name'': | ||
| + | |pBAD strong | ||
| + | |- | ||
| + | |width="60px"|''Input'': | ||
| + | |width="100px"|[http://openwetware.org/wiki/Arabinose L-arabinose] | ||
| + | |- | ||
| + | |''Output'': | ||
| + | | PoPS | ||
| + | |} | ||
| + | </div> | ||
| + | <hr width="800px"> | ||
| + | <div class="noprint" style="padding: 10px; color: #ffffff; background-color: #C0C0C0; width: 800px; align: center"> | ||
| + | <center> | ||
| + | [[Part:BBa_K206000 |Part Main Page]] | ||
| + | [[Part:BBa_K206000:Design |Part Design]] | ||
| + | [[Part:BBa_K206000:Characterization |Characterization]] | ||
| + | [[pBAD Promoter Family |Family]] | ||
| + | [[Part:BBa_K206000:Experience |Add Data]] | ||
| + | </center> | ||
| + | </div> | ||
| + | |} | ||
===Usage and Biology=== | ===Usage and Biology=== | ||
| − | pBAD is an <i>E.coli</i> promoter that is induced by L-arabinose. In the absence of arabinose, the repressor protein AraC (<partinfo>I13458</partinfo>) binds to the AraI1 operator site of pBAD and the upstream operator site AraO2, blocking transcription | + | pBAD is an <i>E.coli</i> promoter that is induced by L-arabinose. In the absence of arabinose, the repressor protein AraC (<partinfo>I13458</partinfo>) binds to the AraI1 operator site of pBAD and the upstream operator site AraO2, blocking transcription [[Part:BBa_K206000#References|[1]]]. In the presence of arabinose, AraC binds to it and changes its conformation such that it interacts with the AraI1 and AraI2 operator sites, permitting transcription [[Part:BBa_K206000#References|[1]]]. |
| + | <br> | ||
| + | <br> | ||
| + | K206000 is a variant pBAD promoter with a modified AraI1 site that has been shown both to be responsive to lower concentrations of arabinose and to exhibit a higher maximum expression than the wild type (<partinfo>I13453</partinfo>), as measured by [[Part:BBa_K206002|coupling to a fluorescent reporter]]. | ||
| + | <br> | ||
| + | <br> | ||
| + | ''What you can do with it:'' | ||
| + | <br> | ||
| + | At a given level of arabinose input, <partinfo>K206000</partinfo> will provide a higher level of PoPS output than its [[pBAD Promoter Family|family members]], allowing analog device responses. See [http://2009.igem.org/Team:British_Columbia our wiki] for a project that makes use of this property. | ||
| + | <br> | ||
| + | <br> | ||
| + | ''Compatibility'': | ||
| + | <br> | ||
| + | Chassis: Best used in the ''E. coli'' strain [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783], which has been modified to permit homogeneous pBAD promoter expression by substituting the chromosomal arabinose-dependent promoter of AraE (arabinose transporter protein) with a constitutive promoter [[Part:BBa_K206000#References|[2]]]. | ||
| + | <br> | ||
| + | Backbone: Has been shown to work on plasmid <partinfo>pSB1C3</partinfo>. | ||
| + | <br> | ||
| + | Reporter: Has been shown to work with reporters <partinfo>I13507</partinfo> and <partinfo>I763020</partinfo>. | ||
| − | [ | + | <br> |
| + | ===<span class='h3bb'>Sequence and Features</span>=== | ||
| + | <partinfo>BBa_K206000 SequenceAndFeatures</partinfo> | ||
| + | <br> | ||
| + | ===References=== | ||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/11102706 [1]] Schlief, R. (2000). Regulation of the L-arabinose operon of ''Escherichia coli''. Trends in Genetics. '''16'''(12):559-565. | ||
| + | <br> | ||
| + | [http://www.ncbi.nlm.nih.gov/pubmed/11739756 [2]] Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, and Keasling JD. (2001). Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. '''147'''(12):3241-7. | ||
| − | + | <!-- Uncomment this to enable Functional Parameter display | |
| + | ===Functional Parameters=== | ||
| + | <partinfo>BBa_K206000 parameters</partinfo> | ||
| + | <!-- --> | ||
| − | + | == Characterized by BNU-China 2019 == | |
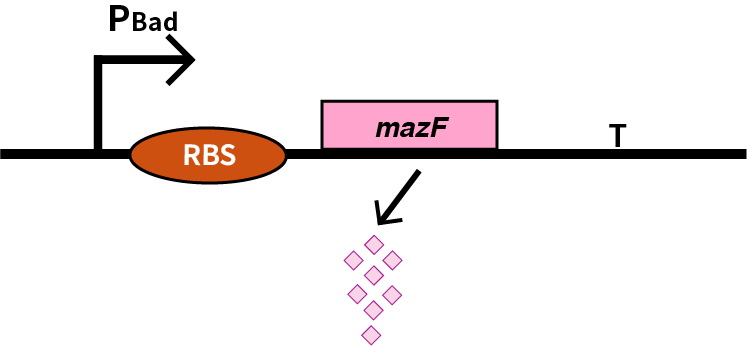
| + | We characterize pBAD (BBa_K206000) by an induced suicide system, in which pBAD controls the downstream mazF (BBa_K302033) gene that serves as a reporter, which encodes an endoribonuclease that cleaves RNAs at ACA sites and causes the death of microbe [1]. As a result, we can characterize pBAD in a cell density-dependent manner in Escherichia coli K-12. | ||
| + | |||
| + | [[Image:2019 BNU-China BBa K3036005 change.png| border | center | 400px]]<br> | ||
| + | |||
| + | In order to characterize pBAD induced by L-arabinose under different concentrations, we take engineered microbe without induction as control group. | ||
| + | |||
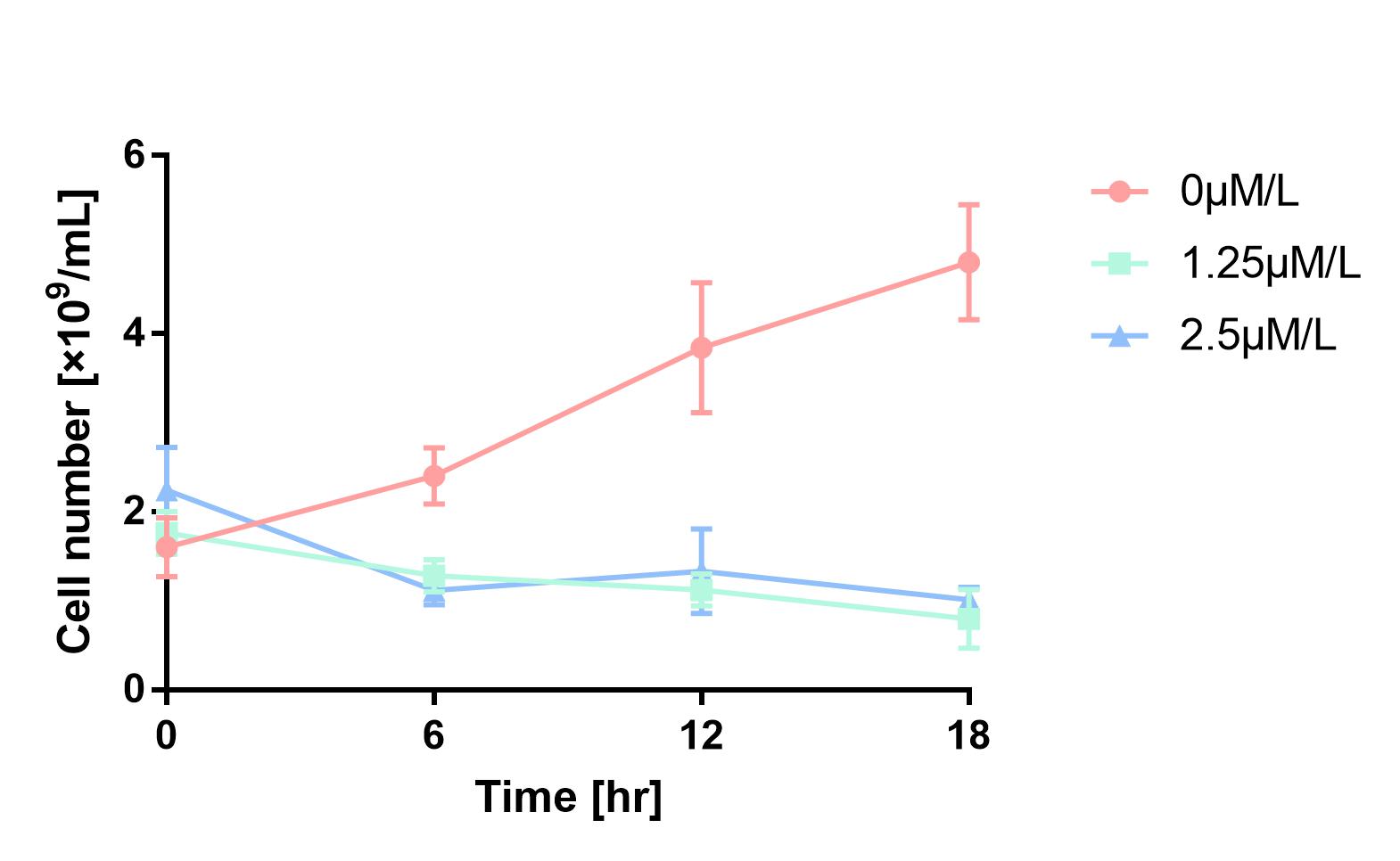
| + | As is shown in Fig.1, the cell number of experimental groups show a significant decrease, which indicates pBAD can be induced by 1.25μM/L and 2.5μM/L arabinose, and 2.5μM/L can be considered as a more effective concentration. | ||
| + | |||
| + | [[Image:2019 BNU-China BBa K302033 change.jpg| border | center | 400px]]<br> | ||
| + | |||
| + | Figure 1 Cell number declines after induction by L-arabinose. It proves that pBAD is induced by L-arabinose. | ||
| + | |||
| + | Beyond our project, pBAD is applied for heterologous gene expression due to its advantages, including moderately high expression levels, induction by a low-cost and non-toxic monosaccharide L-arabinose and tight regulation of transcription, which is particularly significant to expressing toxins. [2] | ||
| + | |||
| + | <b>Experimental approach</b> | ||
| + | |||
| + | 1. Transform the plasmids into E. coli DH5α competent cells. | ||
| + | 2. The engineered bacteria are cultured in 200mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm; | ||
| + | 3. Equally divide the culture into 90 centrifuge tubes, which is 1mL respectively. Centrifuge them at 4000rpm for 5 minutes. Discard the liquid. | ||
| + | 4. Resuspend 30 tubes of collected bacteria with LB-ampicillin (50 ng/µl) containing 1.25μM/L and 2.5μM/L L-arabinose respectively as experimental groups. Resuspend 30 tubes of bacteria with pure LB-ampicillin (50 ng/µl) medium. | ||
| + | 5. Collect 3 tubes of all groups every 6 hours, dilute all of the samples to 107 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately. At the same time, refresh the medium to maintain the concentration of L-arabinose. | ||
| + | 6. Count the number of colonies in 5 cm2 per plate after cultured for 24 hours at 37℃ | ||
| + | 7. Three repicas are tested in each group. | ||
| + | |||
| + | <b>Reference</b> | ||
| + | |||
| + | [1] Nigam A, Ziv T, Oron-Gottesman A, Engelberg-Kulka H2019. Stress-induced MazF-mediated proteins in Escherichia coli. mBio 10: e00340-19. doi:10.1128/mBio.00340-19. | ||
| + | [2] Diana Széliová, Ján Krahulec, Martin Šafránek, et al. Modulation of heterologous expression from PBAD promoter in Escherichia coli production strains[J]. Journal of Biotechnology, 2016, 236:1-9. | ||
| + | |||
| + | |||
| + | <!-- Add more about the biology of this part here | ||
| + | ===Usage and Biology=== | ||
| + | |||
| + | <!-- --> | ||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| − | <partinfo> | + | <partinfo>BBa_K654059 SequenceAndFeatures</partinfo> |
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
| − | <partinfo> | + | <partinfo>BBa_K654059 parameters</partinfo> |
<!-- --> | <!-- --> | ||
Latest revision as of 14:17, 16 October 2019
|
Usage and Biology
pBAD is an E.coli promoter that is induced by L-arabinose. In the absence of arabinose, the repressor protein AraC (BBa_I13458) binds to the AraI1 operator site of pBAD and the upstream operator site AraO2, blocking transcription [1]. In the presence of arabinose, AraC binds to it and changes its conformation such that it interacts with the AraI1 and AraI2 operator sites, permitting transcription [1].
K206000 is a variant pBAD promoter with a modified AraI1 site that has been shown both to be responsive to lower concentrations of arabinose and to exhibit a higher maximum expression than the wild type (BBa_I13453), as measured by coupling to a fluorescent reporter.
What you can do with it:
At a given level of arabinose input, BBa_K206000 will provide a higher level of PoPS output than its family members, allowing analog device responses. See [http://2009.igem.org/Team:British_Columbia our wiki] for a project that makes use of this property.
Compatibility:
Chassis: Best used in the E. coli strain [http://cgsc.biology.yale.edu/Strain.php?ID=111773 BW27783], which has been modified to permit homogeneous pBAD promoter expression by substituting the chromosomal arabinose-dependent promoter of AraE (arabinose transporter protein) with a constitutive promoter [2].
Backbone: Has been shown to work on plasmid pSB1C3.
Reporter: Has been shown to work with reporters BBa_I13507 and BBa_I763020.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 125
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 65
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
References
[http://www.ncbi.nlm.nih.gov/pubmed/11102706 [1]] Schlief, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics. 16(12):559-565.
[http://www.ncbi.nlm.nih.gov/pubmed/11739756 [2]] Khlebnikov A, Datsenko KA, Skaug T, Wanner BL, and Keasling JD. (2001). Homogeneous expression of the PBAD promoter in Escherichia coli by constitutive expression of the low-affinity high-capacity AraE transporter. Microbiology. 147(12):3241-7.
Characterized by BNU-China 2019
We characterize pBAD (BBa_K206000) by an induced suicide system, in which pBAD controls the downstream mazF (BBa_K302033) gene that serves as a reporter, which encodes an endoribonuclease that cleaves RNAs at ACA sites and causes the death of microbe [1]. As a result, we can characterize pBAD in a cell density-dependent manner in Escherichia coli K-12.
In order to characterize pBAD induced by L-arabinose under different concentrations, we take engineered microbe without induction as control group.
As is shown in Fig.1, the cell number of experimental groups show a significant decrease, which indicates pBAD can be induced by 1.25μM/L and 2.5μM/L arabinose, and 2.5μM/L can be considered as a more effective concentration.
Figure 1 Cell number declines after induction by L-arabinose. It proves that pBAD is induced by L-arabinose.
Beyond our project, pBAD is applied for heterologous gene expression due to its advantages, including moderately high expression levels, induction by a low-cost and non-toxic monosaccharide L-arabinose and tight regulation of transcription, which is particularly significant to expressing toxins. [2]
Experimental approach
1. Transform the plasmids into E. coli DH5α competent cells. 2. The engineered bacteria are cultured in 200mL LB-ampicillin (50 ng/µl) medium overnight at 37℃, 200rpm; 3. Equally divide the culture into 90 centrifuge tubes, which is 1mL respectively. Centrifuge them at 4000rpm for 5 minutes. Discard the liquid. 4. Resuspend 30 tubes of collected bacteria with LB-ampicillin (50 ng/µl) containing 1.25μM/L and 2.5μM/L L-arabinose respectively as experimental groups. Resuspend 30 tubes of bacteria with pure LB-ampicillin (50 ng/µl) medium. 5. Collect 3 tubes of all groups every 6 hours, dilute all of the samples to 107 times and then spread them on solid LB-ampicillin (50 ng/µl) medium separately. At the same time, refresh the medium to maintain the concentration of L-arabinose. 6. Count the number of colonies in 5 cm2 per plate after cultured for 24 hours at 37℃ 7. Three repicas are tested in each group.
Reference
[1] Nigam A, Ziv T, Oron-Gottesman A, Engelberg-Kulka H2019. Stress-induced MazF-mediated proteins in Escherichia coli. mBio 10: e00340-19. doi:10.1128/mBio.00340-19. [2] Diana Széliová, Ján Krahulec, Martin Šafránek, et al. Modulation of heterologous expression from PBAD promoter in Escherichia coli production strains[J]. Journal of Biotechnology, 2016, 236:1-9.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]