Difference between revisions of "Part:BBa K1980002"
(→Experience) |
|||
| (19 intermediate revisions by 3 users not shown) | |||
| Line 2: | Line 2: | ||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K1980002 short</partinfo> | <partinfo>BBa_K1980002 short</partinfo> | ||
| + | ==Description== | ||
| + | MymT is a small prokaryotic copper metallothein discovered in <i>Mycobacterium tuberculosis</i>. It is believed that the protein may help the bacterium survive copper toxicity, though deleting the gene had no effect on pathogenicity in mice. It can bind up to 7 copper ions but has a preference for 4-6. | ||
| − | + | A form of MymT was already in the registry at the start of our project <html>(<a href="https://parts.igem.org/Part:BBa_K190020">BBa_K190020</a>) but it was not codon optimised to <i>E. coli</i> and we saw no evidence that it had been used. Here we have codon optimised it and added a C terminal his tag for purification. In part (<a href="https://parts.igem.org/Part:BBa_K1980003">BBa_K1980003</a>) we added a C terminal sfGFP to show if the protein is expressed in E. coli and obtained provisional data, using fluorescence lifetime microscopy, suggesting copper-binding <i>in vivo</i>.</html> | |
| − | + | ||
| − | A form of MymT was already in the registry at the start of our project (https://parts.igem.org/Part:BBa_K190020) but it was | + | |
<!-- --> | <!-- --> | ||
| Line 27: | Line 27: | ||
==Experience== | ==Experience== | ||
<p> We cloned MymT from Gblock into the shipping vector then transferred it into the pBAD, arabinose-inducible commercial expression vector.</p> | <p> We cloned MymT from Gblock into the shipping vector then transferred it into the pBAD, arabinose-inducible commercial expression vector.</p> | ||
| − | + | ||
| + | ===Absorbance Assay=== | ||
| + | |||
<p>We were unable however to detect copper chelation activity of MymT when expressed from pBAD in MG1655 <i>E. coli</i> strain, using an absorbance assay. | <p>We were unable however to detect copper chelation activity of MymT when expressed from pBAD in MG1655 <i>E. coli</i> strain, using an absorbance assay. | ||
</p> | </p> | ||
<p> | <p> | ||
| − | The assay used was the reagent Bathocuproine disulphonic Acid (BCS). BCS is colourless in the absence of Cu+ but upon exposure to Cu+, BCS forms complexes with Cu+ and absorbs strongly in the 480nm range. In our assays, at concentrations of 50µg/mL, the 480nm absorbance varied linearly with the Cu+ concentration from the detection limit of around 1µM Cu+, to approximately 20µM Cu+. As not only MymT and Csp1 bind copper in the Cu+ form, but the assay also requires singly charged copper, the assays were | + | The assay used was the reagent Bathocuproine disulphonic Acid (BCS). BCS is colourless in the absence of Cu<sup>+</sup> but upon exposure to Cu<sup>+</sup>, BCS forms complexes with Cu<sup>+</sup> and absorbs strongly in the 480nm range. In our assays, at concentrations of 50µg/mL, the 480nm absorbance varied linearly with the Cu+ concentration from the detection limit of around 1µM Cu<sup>+</sup>, to approximately 20µM Cu<sup>+</sup>. As not only MymT and Csp1 bind copper in the Cu<sup>+</sup> form, but the assay also requires singly charged copper, the assays were optimised to include a suitable about of mild reducing agent to ensure reduction of the added CuSO<sub>4</sub> (releases Cu<sup>2+</sup>) to Cu<sup>+</sup>. After trying L(+)-Ascorbate and DL-Dithiothreitol, and L-Glutathione as candidates, L-Glutathione was selected as it was both mild enough to not damage biological material and efficient at reducing Cu<sup>2+</sup>. At >2-3 times the concentration of Cu<sup>2+</sup> in solution, L-Glutathione had maximum reductive activity against Cu<sup>2+</sup>.</p> |
| − | </p> Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount needed to be detectable on the assay ( | + | <p> Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount needed to be detectable on the assay (1μM detection limit).</p> |
| − | <p>We purified a sfGFP-tagged version of this protein (BBa K1980003) but were still unable to detect copper chelation with the assay in our purified extracts.</p> | + | <p>We purified a sfGFP-tagged version of this protein <html>(<a href="https://parts.igem.org/Part:BBa_K1980003">BBa K1980003</a>)</html> but were still unable to detect copper chelation with the assay in our purified extracts.</p> |
| + | |||
===FLIM=== | ===FLIM=== | ||
<p> | <p> | ||
| Line 62: | Line 65: | ||
<p> | <p> | ||
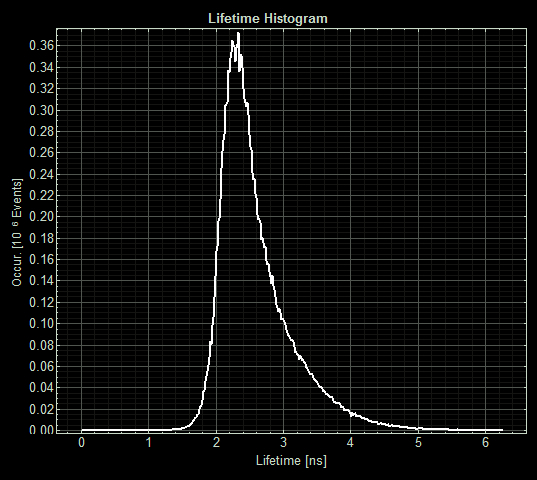
When MymTsfGFP was induced the mean lifetime decreased to 2.3ns. As MymTsfGFP is was observed to be reliably expressed and because MymT is a small copper cluster separated form sfGFP by a small linker we believe that this represents additional quenching of the fluorphore by MymT-bound copper showing <i>in vivo</i> copper chelation.</p> | When MymTsfGFP was induced the mean lifetime decreased to 2.3ns. As MymTsfGFP is was observed to be reliably expressed and because MymT is a small copper cluster separated form sfGFP by a small linker we believe that this represents additional quenching of the fluorphore by MymT-bound copper showing <i>in vivo</i> copper chelation.</p> | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
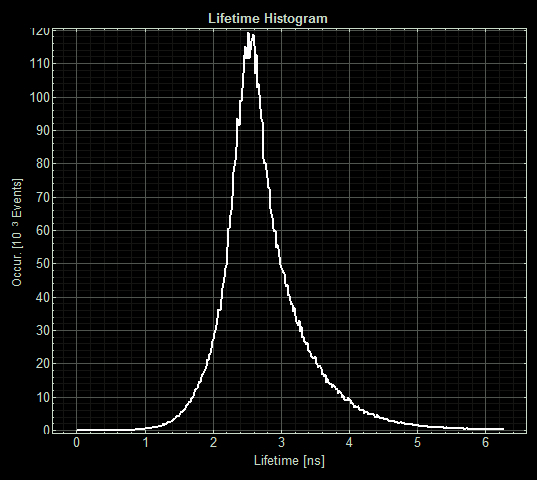
| + | [[File:T--Oxford--Sam-FLIM-sfGFP.jpg|400px|thumb|center|Histogram showing the fluorescence lifetime of sfGFP at 5uM copper]] | ||
| + | [[File:T--Oxford--Sam-FLIM-MymTsfGFP.jpg|400px|thumb|center|Histogram showing the fluorescence lifetime of Csp1sfGFP at 5uM copper]] | ||
| Line 108: | Line 81: | ||
<p style="text-align: right"><i>Author: Sam Garforth and Andreas Hadjicharalambous</i></p> | <p style="text-align: right"><i>Author: Sam Garforth and Andreas Hadjicharalambous</i></p> | ||
| + | |||
| + | ==Characterisation by 2019 Hong Kong JSS Team== | ||
| + | Group: iGEM 2019 Hong Kong JSS (2019-10-11) | ||
| + | <p>Author: Jamie Tam</p> | ||
| + | |||
| + | ===MyMT expressing <i>E. coli</i> showed enchanced copper absorption=== | ||
| + | |||
| + | Our project aimed to use <i>E. coli</i> as the metal pollutant absorbent in liquid medium. We cloned and transformed different metallothionein genes into <i>E. coli</i> to study the effect on copper removal ability in the organism. | ||
| + | This part <html>(<a href="https://parts.igem.org/Part:BBa_K1980002">BBa K1980002</a>)</html> was expressed under the regulations of strong constitutive promoter <html>(<a href="https://parts.igem.org/Part:BBa_J23100">BBa J23100</a>)</html>, RBS <html>(<a href="https://parts.igem.org/Part:BBa_B0032">BBa B0032</a>)</html> and double terminator <html>(<a href="https://parts.igem.org/Part:BBa_B0015">BBa B0015</a>)</html>. | ||
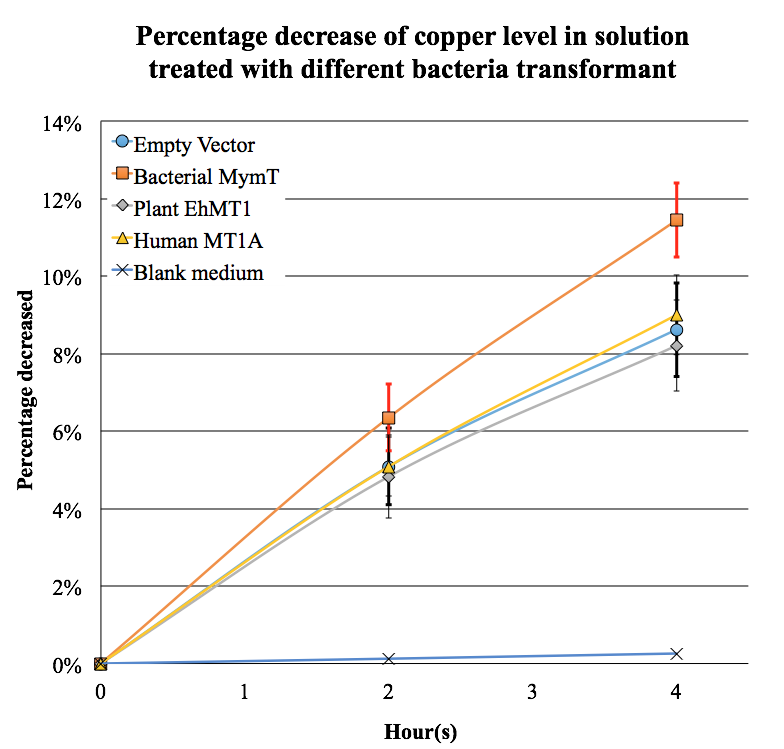
| + | <p>The result showed that the copper (II) ion concentration in the medium decreased by about 12% when treated with <i>MyMT</i>expressing <i>E. coli</i>. Meanwhile, the other <i>MT</i> transformant groups and the control groups showed only ~8% of decrease. </p> | ||
| + | <p> | ||
| + | [[File:BBa K1980002 copper removal.png|400px|thumb|center|The decrease of copper concentration in <i>MyMT</i> transformed group was significant when compared with control groups after 4 hours of incubation]]</p> | ||
| + | <p> | ||
| + | Although there was an improvement in the removal effect, we seek to explore more <i>MTs</i> that have the potential to further enhance our project. We also studied metallothionein from <i>Corynebacterium glutamicum</i> <html>(<a href="https://parts.igem.org/Part:BBa_K3076100">BBa K3076100</a>)</html>.and found the results were more satisfying when compared with <i>MyMT</i>. </p> | ||
| + | |||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
Latest revision as of 12:43, 13 October 2019
MymT
Description
MymT is a small prokaryotic copper metallothein discovered in Mycobacterium tuberculosis. It is believed that the protein may help the bacterium survive copper toxicity, though deleting the gene had no effect on pathogenicity in mice. It can bind up to 7 copper ions but has a preference for 4-6.
A form of MymT was already in the registry at the start of our project (BBa_K190020) but it was not codon optimised to E. coli and we saw no evidence that it had been used. Here we have codon optimised it and added a C terminal his tag for purification. In part (BBa_K1980003) we added a C terminal sfGFP to show if the protein is expressed in E. coli and obtained provisional data, using fluorescence lifetime microscopy, suggesting copper-binding in vivo.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Usage and Biology
Our project aimed to detect and chelate dietary copper as a treatment for Wilson's Disease, a copper accumulation disorder. We decided that the ideal copper chelation protein would have these properties:
- Should be able to bind multiple copper ions per peptide to increase the efficient use of cell resources.
- They should be from the prokaryotic domain because eukaryotic proteins can have expression issues in Escherichia coli.
MymT is a small prokaryotic metallothein discovered in Mycobacterium tuberculosis by Gold et al.(1). It is believed that the protein may help the bacterium survive copper toxicity.
MymT is believed to bind up to 7 copper ions but has a preference for 4-6.(1)
Experience
We cloned MymT from Gblock into the shipping vector then transferred it into the pBAD, arabinose-inducible commercial expression vector.
Absorbance Assay
We were unable however to detect copper chelation activity of MymT when expressed from pBAD in MG1655 E. coli strain, using an absorbance assay.
The assay used was the reagent Bathocuproine disulphonic Acid (BCS). BCS is colourless in the absence of Cu+ but upon exposure to Cu+, BCS forms complexes with Cu+ and absorbs strongly in the 480nm range. In our assays, at concentrations of 50µg/mL, the 480nm absorbance varied linearly with the Cu+ concentration from the detection limit of around 1µM Cu+, to approximately 20µM Cu+. As not only MymT and Csp1 bind copper in the Cu+ form, but the assay also requires singly charged copper, the assays were optimised to include a suitable about of mild reducing agent to ensure reduction of the added CuSO4 (releases Cu2+) to Cu+. After trying L(+)-Ascorbate and DL-Dithiothreitol, and L-Glutathione as candidates, L-Glutathione was selected as it was both mild enough to not damage biological material and efficient at reducing Cu2+. At >2-3 times the concentration of Cu2+ in solution, L-Glutathione had maximum reductive activity against Cu2+.
Modelling by our team suggested that this was because insufficient protein could be expressed to chelate the amount needed to be detectable on the assay (1μM detection limit).
We purified a sfGFP-tagged version of this protein (BBa K1980003) but were still unable to detect copper chelation with the assay in our purified extracts.
FLIM
We discovered a paper by Hötzer et al(2) that described how His-tagged GFP can be quenched by a copper ion binding to this His tag leading to a reduction in the fluorescence lifetime (the time the fluorophore spends in the excited state before returning to the ground state by emitting a photon.) They speculated that this could potentially be used as a in vivo copper assay.

As we had His-tagged our chelator-sfGFP constructs we were curious to see if this technique could be applied to our parts to measure copper chelation in vivo by our parts. We believe that two possibilities were likely:
- Copper chelation by the chelator reduces the free copper concentration inside the cell meaning that less binds to the His tag and the fluorescence lifetime will be greater than a His-tagged sfGFP control
- Copper chelation by the chelator would allow additional quenching if copper was bound within the quenching radius of the fluorophore leading to a reduction in fluorescence lifetime compare with a sfGFP control
Lacking access to a fluorescence lifetime microscope ourselves we contacted Cardiff iGEM who had a FLIM machine in their bioimaging unit.
We sent Cardiff iGEM our parts MymTsfGFP in pBAD and pCopA CueR sfGFP (as a control) in live MG1655 E. coli in agar tubes. Cardiff grew them overnight in 5ml of LB with 5uM copper with and without 2mM arabinose.
The imaging unit spread each strain on slides and measured the fluorescence lifetime of three areas on each slide.
(Acquisition parameters: using the x63 water immersion objective with excitation at 483nm (71% intensity, pulse rate 40MHz) and emission via a BP500-550 filter. Scan resolution at 512 x 512 pixels at pixel size of 0.26 microns/pixel, 1AU pinhole. Counts of >1000 per lifetime recording.)
FLIM images from one section of each slide:

As expected the pCopA CueR sfGFP control was fluorescent, with and without arabinose, with the mean fluorescence lifetime a consistent 2.6ns.
When MymTsfGFP was induced the mean lifetime decreased to 2.3ns. As MymTsfGFP is was observed to be reliably expressed and because MymT is a small copper cluster separated form sfGFP by a small linker we believe that this represents additional quenching of the fluorphore by MymT-bound copper showing in vivo copper chelation.
References
(1) Ben Gold, Haiteng Deng, Ruslana Bryk, Diana Vargas, David Eliezer, Julia Roberts, Xiuju Jiang, & Carl Nathan (2009) “Identification of a Copper-Binding Metallothionein in Pathogenic Mycobacteria” Nat Chem Biol. 2008 October ; 4(10): 609–616. doi:10.1038/nchembio.109.
(2) Hötzer B., Ivanov R., Altmeier S., Kappl R., Jung G., (2011) "Determination of copper(II) ion concentration by lifetime measurements of green fluorescent protein." Journal of Fluorescence, 21(6), pp. 2143-2153. doi: 10.1007/s10895-011-0916-1
Author: Sam Garforth and Andreas Hadjicharalambous
Characterisation by 2019 Hong Kong JSS Team
Group: iGEM 2019 Hong Kong JSS (2019-10-11)
Author: Jamie Tam
MyMT expressing E. coli showed enchanced copper absorption
Our project aimed to use E. coli as the metal pollutant absorbent in liquid medium. We cloned and transformed different metallothionein genes into E. coli to study the effect on copper removal ability in the organism. This part (BBa K1980002) was expressed under the regulations of strong constitutive promoter (BBa J23100), RBS (BBa B0032) and double terminator (BBa B0015).
The result showed that the copper (II) ion concentration in the medium decreased by about 12% when treated with MyMTexpressing E. coli. Meanwhile, the other MT transformant groups and the control groups showed only ~8% of decrease.
Although there was an improvement in the removal effect, we seek to explore more MTs that have the potential to further enhance our project. We also studied metallothionein from Corynebacterium glutamicum (BBa K3076100).and found the results were more satisfying when compared with MyMT.