Difference between revisions of "Part:BBa K2920727"
| Line 31: | Line 31: | ||
[[File:T--Tunghai TAPG--gfp2.png]] | [[File:T--Tunghai TAPG--gfp2.png]] | ||
Figure.3 Ex/Em=488/518 nm normalized with OD600 | Figure.3 Ex/Em=488/518 nm normalized with OD600 | ||
| + | ------------------------------------------------------ | ||
| + | |||
| + | the condition of the characterization | ||
| − | |||
Day 1 ↓ culture E. coli BL21(DE3) carrying vector only or T7-RBS-EGFP in LB + Kan (50 ug/ml) O/N at 37°C | Day 1 ↓ culture E. coli BL21(DE3) carrying vector only or T7-RBS-EGFP in LB + Kan (50 ug/ml) O/N at 37°C | ||
Revision as of 10:51, 13 October 2019
Effective AMP (Antimicrobial Peptide)
BBa_K2920727 antimicrobial peptide is our target gene from 2019 Tunghai_TAPG iGEM team, we can prove the antimicrobial function of the peptide after we translated our peptide from the amino acid to the DNA code, so we mass production our specific peptide by genetic engineering method and made it into the product. AMPs is a currently a potential defense against multidrug-resistant (MDR) pathogens which is naturally occurring molecular as an alternative to antibiotic. This kind of molecular have broad activity to directly kill many organisms for instance bacteria, yeast, and fungi etc. Peptide is composed of different length and different arrangement of amino acid. Unlike antibiotic, AMPs have diverse mechanism against bacteria which make it hard to develop a complete resistance. Generally, bacteria membrane is composed of liquid bilayer structure. TungHai iGEM aim to ameliorate the hospital environment by air purifier(the antimicrobial peptide liquid), we observed that people are suffering HAI (Hospital Acquired Infection) these days. HAI (Hospital acquired infection) literally it is an infection get caught during hospitalization. To be more academically, we call it nosocomial in medical. Most of the time, HAI is often cause by the bacteria since recently the increase of the abuse of antibiotic in the hospital
<Usage>
TungHai iGEM aim to ameliorate the hospital environment by air purifier(the antimicrobial peptide liquid), we observed that people are suffering HAI (Hospital Acquired Infection) these days. HAI (Hospital acquired infection) literally it is an infection get caught during hospitalization. To be more academically, we call it nosocomial in medical. Most of the time, HAI is often cause by the bacteria since recently the increase of the abuse of antibiotic in the hospital
<Biology>
<Characterization> Characterization of Fluorescence intensity
The TAPG tunghai team 2019 are using our peptide jj01’s back end combined with BBa_K1911005, and adding IPTG induction for expression, to ensure the accuracy of our protein expression, and using fluorescent protein as the identification, to get the yield of our protein. Among of these, we have four different cells in total (normal BL21, BL21 with fluorescent protein, BL21 adding IPTG, BL21 with fluorescent protein adding IPTG), we are using these three ways to ensure the accuracy.
In figure 1., the x-axis refers to time, the y-axis refers to OD value. We tested the OD600 of four cells, continually to 24 hours, observed the growth curve. Apparently, there is no obvious gap of these four cells.
In figure 2., the x-axis refers to time, the y-axis refers to Fluorescence intensity. We tested four cells’ Fluorescence intensity individually (Ex/Em=488/518nm), we could see that, the curves of two cells without EGFP are not overlapping, and at the very bottom of the chart. As for the cell with IEGFG but without IPTG induction, the curve stay remain and send out tiny fluorescent. When IPTG are adding in the cell with fluorescent, we could see that the curve has an obvious change, we add 1 mm IPTG, when OD are grown to 0.8, we can know that the curve has a dramatically growth.
In figure 3., the x-axis refers to time, the y-axis refers to Fluorescence intensity. We divided figure 2. by figure 1., get the Fluorescence intensity of four different kind of cells per unit; then we are able to measure our productions correctly by knowing Fluorescence intensity from each cells. Because the quantity of Fluorescence protein was proportional to the target gene that is going to be expressed. In order to know how many Fluorescence intensity each bacteria produces, we have to divide the numbers of bacteria by total Fluorescence intensity, what we get is the quantity of the Fluorescence intensity per bacteria produces.
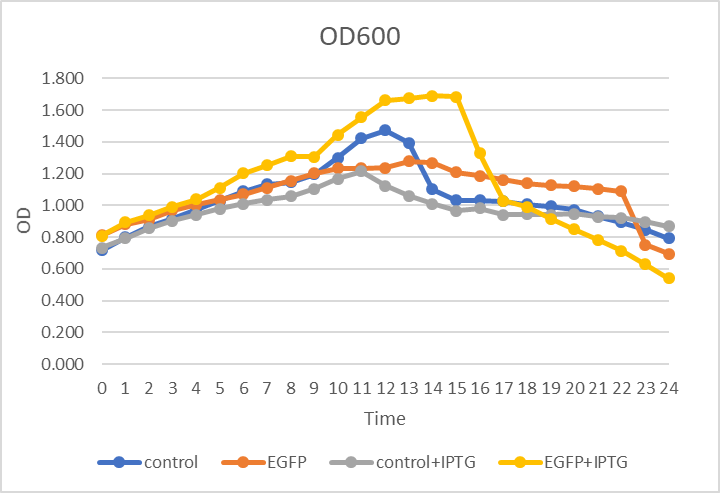 Figure.1 OD600
Figure.1 OD600
 Figure.2 Ex/Em=488/518 nm
Figure.2 Ex/Em=488/518 nm
 Figure.3 Ex/Em=488/518 nm normalized with OD600
Figure.3 Ex/Em=488/518 nm normalized with OD600
the condition of the characterization
Day 1 ↓ culture E. coli BL21(DE3) carrying vector only or T7-RBS-EGFP in LB + Kan (50 ug/ml) O/N at 37°C
Day 2 ↓ measure OD600
↓ dilute to OD600 around 0.1
↓ shake at 200 rpm, 37°C until OD600 around 0.8
↓ add 1mM IPTG for protein induction
↓ measure OD600 and Ex/Em=488/518 nm every 30min for 24hr
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
