Difference between revisions of "Part:BBa J23102"
| Line 29: | Line 29: | ||
<p>We wanted to add new, normalised RFU data for two promoters from the Anderson family of constitutive promoters BBa_J23102 (Strong constitutive promoter) and BBa_J23106 (weak constitutive promoter) respectively; and a T7 promoter BBa_K199118 all expressing mRFP1. Another interest that we had was to test how OD was affected by RFP production, therefore we performed our measurements twice, at OD600 and OD660 respectively. This was done because it has been shown that OD660 gives a more accurate representation of bacterial growth in RFP-producing bacteria (Hecht et al., 2016). </p> | <p>We wanted to add new, normalised RFU data for two promoters from the Anderson family of constitutive promoters BBa_J23102 (Strong constitutive promoter) and BBa_J23106 (weak constitutive promoter) respectively; and a T7 promoter BBa_K199118 all expressing mRFP1. Another interest that we had was to test how OD was affected by RFP production, therefore we performed our measurements twice, at OD600 and OD660 respectively. This was done because it has been shown that OD660 gives a more accurate representation of bacterial growth in RFP-producing bacteria (Hecht et al., 2016). </p> | ||
<p>In order to obtain our results, we grew our cell cultures up to an OD600/660 of ~0.6. After the desired OD had been reached, cultures were induced with IPTG (for BBa_K199118) and anhydrotetracycline (for BBa_K092300). Then they were placed on a microplate reader (CLARIOstar®, BMG Labtech). The specific script conditions can be seen below. The machine was set to measure OD600/660 and RFU overnight every 15 minutes. The data was then analysed and plotted. This experiment was performed both in transformed E. coli DH5a as well as BL21.</p> | <p>In order to obtain our results, we grew our cell cultures up to an OD600/660 of ~0.6. After the desired OD had been reached, cultures were induced with IPTG (for BBa_K199118) and anhydrotetracycline (for BBa_K092300). Then they were placed on a microplate reader (CLARIOstar®, BMG Labtech). The specific script conditions can be seen below. The machine was set to measure OD600/660 and RFU overnight every 15 minutes. The data was then analysed and plotted. This experiment was performed both in transformed E. coli DH5a as well as BL21.</p> | ||
| − | <p style="font-size: | + | <p style="font-size: 110%; font-weight: bold; ">OD600:</p> |
<p style="margin: 0!important;">Discrete wavelengths, 1</p> | <p style="margin: 0!important;">Discrete wavelengths, 1</p> | ||
<p style="margin: 0!important;">Wavelength: 600</p> | <p style="margin: 0!important;">Wavelength: 600</p> | ||
<p style="margin: 0!important;">Well scan: spiral average, 5mm diameter</p> | <p style="margin: 0!important;">Well scan: spiral average, 5mm diameter</p> | ||
| − | <p style="font-size: | + | <p style="font-size: 110%; font-weight: bold;">OD660:</p> |
<p style="margin: 0!important;">Discrete wavelengths, 1</p> | <p style="margin: 0!important;">Discrete wavelengths, 1</p> | ||
<p style="margin: 0!important;">Wavelength: 660</p> | <p style="margin: 0!important;">Wavelength: 660</p> | ||
<p style="margin: 0!important;">Well scan: spiral average, 5mm diameter</p> | <p style="margin: 0!important;">Well scan: spiral average, 5mm diameter</p> | ||
| − | <p style="font-size: | + | <p style="font-size: 110%; font-weight: bold;">RFP Fluorescence:</p> |
<p style="margin: 0!important;">Focal Height: 7.5</p> | <p style="margin: 0!important;">Focal Height: 7.5</p> | ||
<p style="margin: 0!important;">Gain: 1000</p> | <p style="margin: 0!important;">Gain: 1000</p> | ||
Revision as of 19:17, 12 October 2019
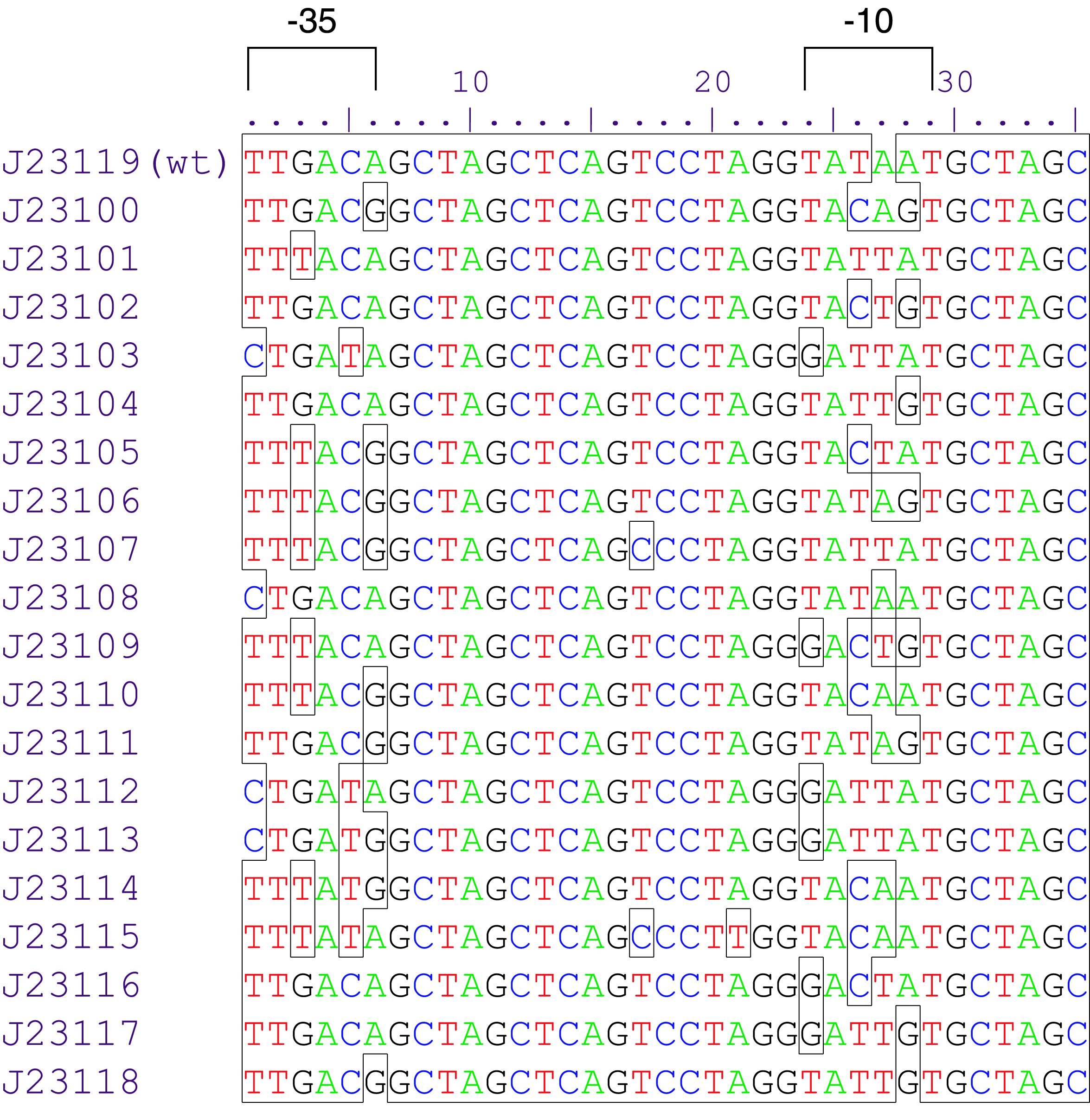
constitutive promoter family member
Variant RFP (au) J23112 1 J23103 17 J23113 21 J23109 106 J23117 162 J23114 256 J23115 387 J23116 396 J23105 623 J23110 844 J23107 908 J23106 1185 J23108 1303 J23118 1429 J23111 1487 J23101 1791 J23104 1831 J23102 2179 J23100 2547 |
Constitutive promoter family
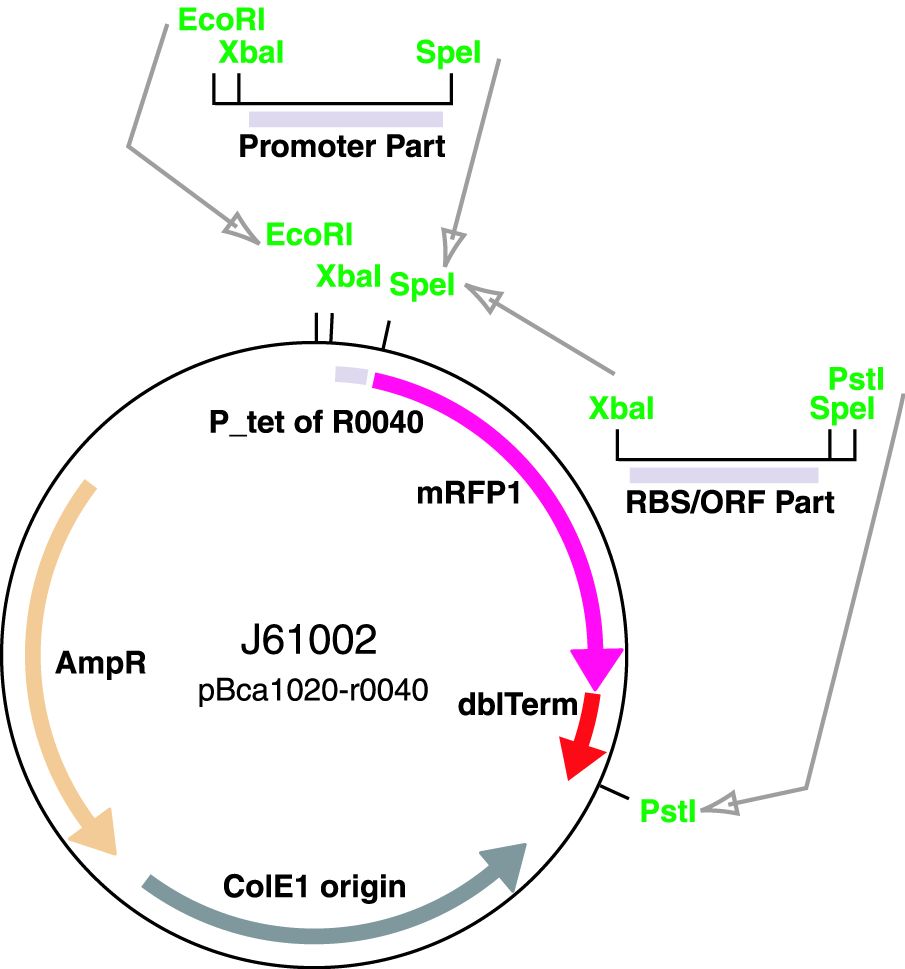
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part BBa_J61002 for details on their use.
These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. JCAraw
Contribution
Group: Valencia_UPV iGEM 2018
Author: Adrián Requena Gutiérrez, Carolina Ropero
Summary: We adapted the part to be able to assemble transcriptional units with the Golden Gate assembly method
Documentation:
In order to create our complete [http://2018.igem.org/Team:Valencia_UPV/Part_Collection part collection] of parts compatible with the Golden Gate assembly method, we made the part BBa_K2656005 which is this part adapted to the Golden Gate technology.
Sequence and Features
- 10INCOMPATIBLE WITH RFC[10]Unknown
- 12INCOMPATIBLE WITH RFC[12]Unknown
- 21INCOMPATIBLE WITH RFC[21]Unknown
- 23INCOMPATIBLE WITH RFC[23]Unknown
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
Manchester 2019 Characterisation:
We wanted to add new, normalised RFU data for two promoters from the Anderson family of constitutive promoters BBa_J23102 (Strong constitutive promoter) and BBa_J23106 (weak constitutive promoter) respectively; and a T7 promoter BBa_K199118 all expressing mRFP1. Another interest that we had was to test how OD was affected by RFP production, therefore we performed our measurements twice, at OD600 and OD660 respectively. This was done because it has been shown that OD660 gives a more accurate representation of bacterial growth in RFP-producing bacteria (Hecht et al., 2016).
In order to obtain our results, we grew our cell cultures up to an OD600/660 of ~0.6. After the desired OD had been reached, cultures were induced with IPTG (for BBa_K199118) and anhydrotetracycline (for BBa_K092300). Then they were placed on a microplate reader (CLARIOstar®, BMG Labtech). The specific script conditions can be seen below. The machine was set to measure OD600/660 and RFU overnight every 15 minutes. The data was then analysed and plotted. This experiment was performed both in transformed E. coli DH5a as well as BL21.
OD600:
Discrete wavelengths, 1
Wavelength: 600
Well scan: spiral average, 5mm diameter
OD660:
Discrete wavelengths, 1
Wavelength: 660
Well scan: spiral average, 5mm diameter
RFP Fluorescence:
Focal Height: 7.5
Gain: 1000
Excitation: 574-15
Emission: 618-22
Well scan: Matrix scan 3x3 1mm diameter