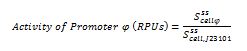Difference between revisions of "Part:BBa J23108:Experience"
| (6 intermediate revisions by 5 users not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | This experience page is provided so that any user may enter their experience using this part.<BR>Please enter | ||
| Line 19: | Line 18: | ||
<!-- End of the user review template --> | <!-- End of the user review template --> | ||
<!-- DON'T DELETE --><partinfo>BBa_J23108 EndReviews</partinfo> | <!-- DON'T DELETE --><partinfo>BBa_J23108 EndReviews</partinfo> | ||
| + | |||
| + | Andrew Kirk, undergraduate, Penn State iGEM 2010 | ||
| + | |||
| + | This constitutive promoter has had inconsistent sequencing results in the 2010 distribution. Our team found that the sequence | ||
| + | |||
| + | TTGATGGCTAGCTCAGTCCTAGGTACNATGCTAGC | ||
| + | |||
| + | consistently came up in place of the intended sequence. It is possible that our distribution is a fluke. Has any other team experienced similar results? | ||
| + | |||
| + | |||
| + | <I>'''iGEM CINVESTAV_IPN_UNAM''' CHARACTERIZATION OF IGEM DISTRIBUTION BIOPARTS</I> | ||
| + | |||
| + | For contribute to the parts registry our team decided to make the characterization of constitutive promoters, in ''E. coli'', belonging to the family isolated from a small combinatorial library (J23101 , J23102, J23104, J23107, J23108, J2311, and J23115) which were attached to GFP, in psB1C3, to determine promoter activity, using the equipment Victor X3 Multilabel Plate Reader. | ||
| + | |||
| + | [[Image:Gfp1.jpg]] | ||
| + | |||
| + | '''Fig. 1 Construction of the promoter J23108 expressing GFP.''' | ||
| + | |||
| + | '''Methods''' | ||
| + | |||
| + | With the selected colonies, an overnight culture was made in M9 media(minimal media supplemented with 0.2% CAA). After 12 hours the culture was transferred to a 96 well plate at a 1:10 dilution (20 μl of culture and 180 μL of fresh M9 medium). | ||
| + | OD and fluorescence measurements of the selected colonies were performed at intervals of 30 minutes for 16 h. | ||
| + | From the results the PopS were calculated (polymerases per second). | ||
| + | |||
| + | '''Modeling''' | ||
| + | |||
| + | The ecuations used for calulated de promoter activity were based on (R. K. Jason et. al 2009). | ||
| + | |||
| + | [[Image:ecu4.jpg]] | ||
| + | |||
| + | [[Image:ecu5.jpg]] | ||
| + | |||
| + | '''Results''' | ||
| + | |||
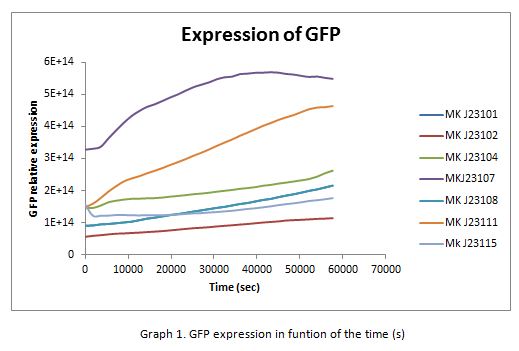
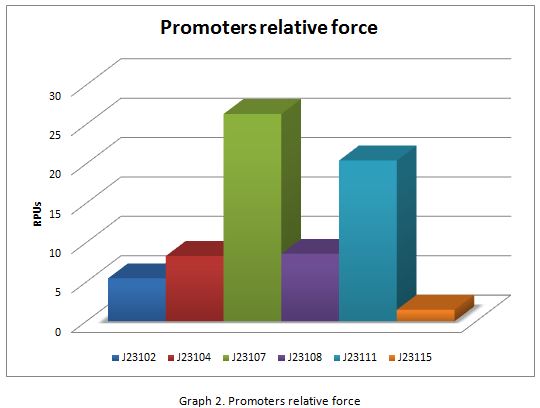
| + | In the following graphs there is shown the GFP expression in function of th time and the realtive promotor intensity. | ||
| + | |||
| + | |||
| + | [[Image:graf1.jpg]] | ||
| + | |||
| + | [[Image:graf2.jpg]] | ||
| + | |||
| + | |||
| + | With the previous results of the characterization of the promoters there is concluded that the promoter J23107, is the strongest because it produces more RPUs” | ||
| + | {|width='80%' style='border:1px solid gray' | ||
| + | |- | ||
| + | |width='10%'| | ||
| + | <partinfo>BBa_J23108 AddReview 3</partinfo> | ||
| + | <I>2017 OUC-China</I> | ||
| + | |width='60%' valign='top'| | ||
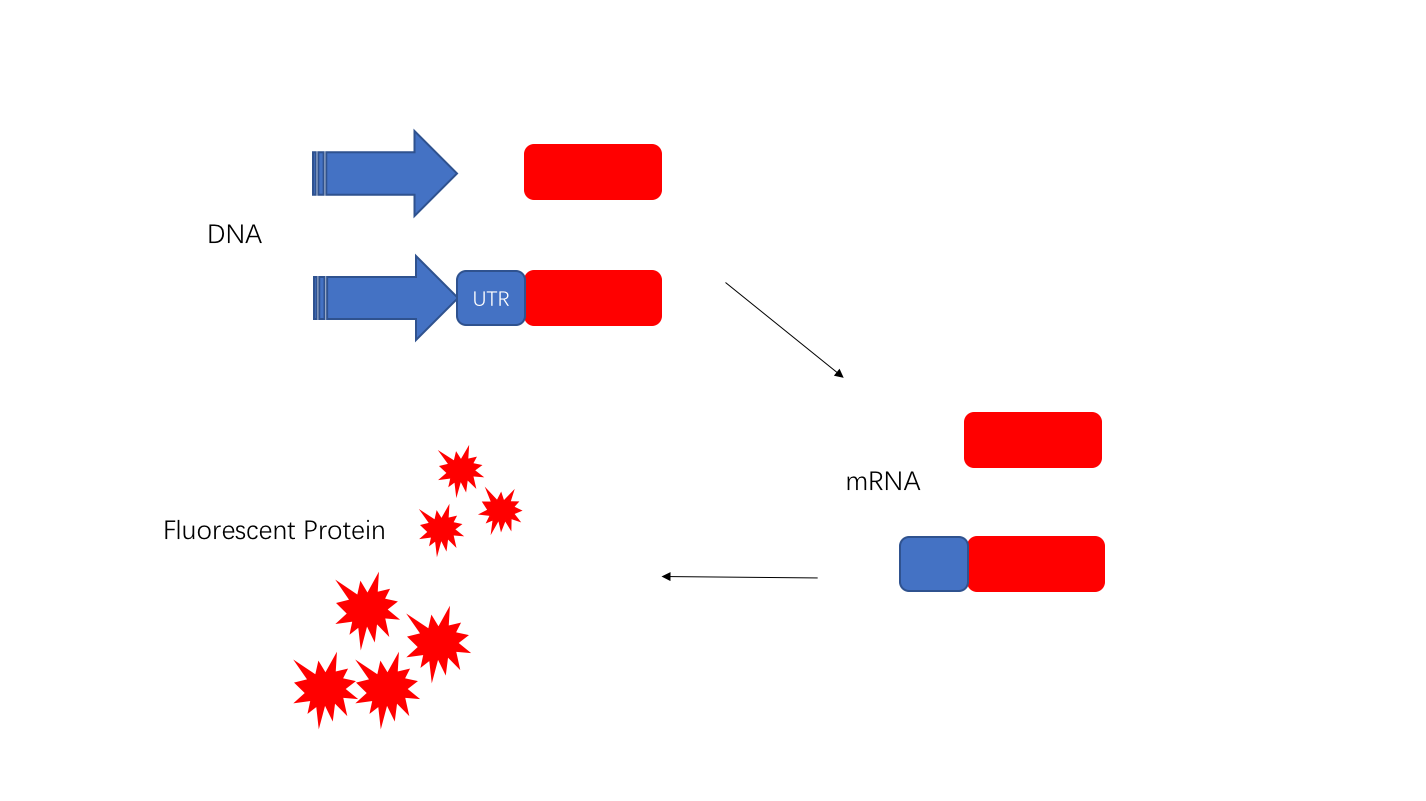
| + | 5'UTR will be transcribed and can be used as a regulatory element to adjust the translation process. We chose J23108 and added the 5'UTR sequence (UTRrpsT)thereafter. We found that 6h fluorescence intensity increased by more than 1.5-fold. | ||
| + | |||
| + | [[Image:T--OUC-China--improve.illustration.png|500px|thumb|center|UTRrpsT enhances protein expression by structure at the 5’UTR.]] | ||
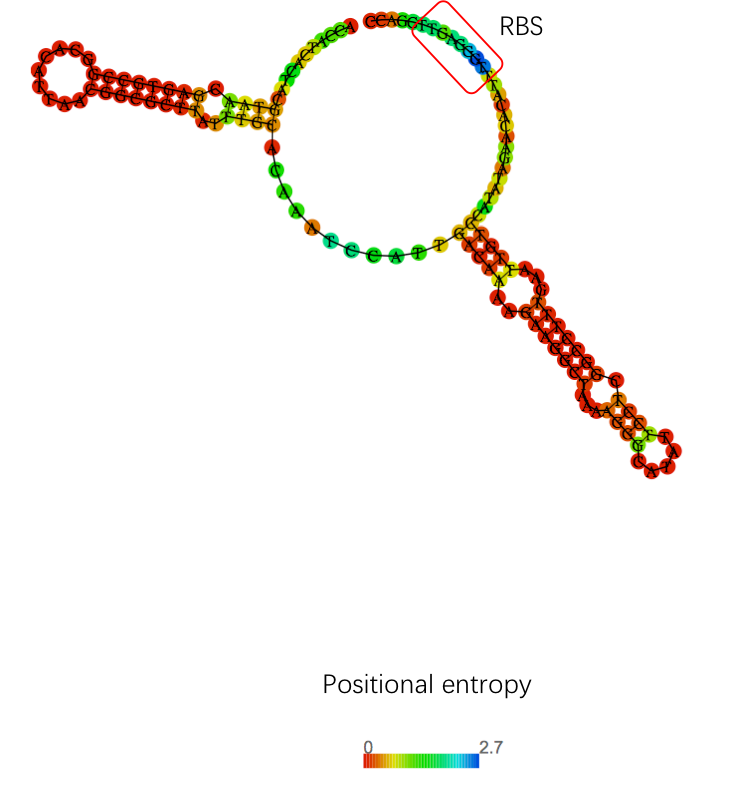
| + | [[Image:T--OUC-China-utrrbs.png|500px|thumb|center|https://static.igem.org/mediawiki/parts/2/25/T--OUC-China-utrrbs.png|The enhancement of UTRrpsT may be related to its secondary structure, as shown by the website (http://rna.tbi.univie.ac.at/).]] | ||
| + | |}; | ||
Latest revision as of 04:22, 10 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_J23108
User Reviews
UNIQc0d6127ca8e6fabd-partinfo-00000000-QINU UNIQc0d6127ca8e6fabd-partinfo-00000001-QINU
Andrew Kirk, undergraduate, Penn State iGEM 2010
This constitutive promoter has had inconsistent sequencing results in the 2010 distribution. Our team found that the sequence
TTGATGGCTAGCTCAGTCCTAGGTACNATGCTAGC
consistently came up in place of the intended sequence. It is possible that our distribution is a fluke. Has any other team experienced similar results?
iGEM CINVESTAV_IPN_UNAM CHARACTERIZATION OF IGEM DISTRIBUTION BIOPARTS
For contribute to the parts registry our team decided to make the characterization of constitutive promoters, in E. coli, belonging to the family isolated from a small combinatorial library (J23101 , J23102, J23104, J23107, J23108, J2311, and J23115) which were attached to GFP, in psB1C3, to determine promoter activity, using the equipment Victor X3 Multilabel Plate Reader.
Fig. 1 Construction of the promoter J23108 expressing GFP.
Methods
With the selected colonies, an overnight culture was made in M9 media(minimal media supplemented with 0.2% CAA). After 12 hours the culture was transferred to a 96 well plate at a 1:10 dilution (20 μl of culture and 180 μL of fresh M9 medium). OD and fluorescence measurements of the selected colonies were performed at intervals of 30 minutes for 16 h. From the results the PopS were calculated (polymerases per second).
Modeling
The ecuations used for calulated de promoter activity were based on (R. K. Jason et. al 2009).
Results
In the following graphs there is shown the GFP expression in function of th time and the realtive promotor intensity.
With the previous results of the characterization of the promoters there is concluded that the promoter J23107, is the strongest because it produces more RPUs”
|
•••
2017 OUC-China |
5'UTR will be transcribed and can be used as a regulatory element to adjust the translation process. We chose J23108 and added the 5'UTR sequence (UTRrpsT)thereafter. We found that 6h fluorescence intensity increased by more than 1.5-fold. |