Difference between revisions of "Part:BBa K1680009:Experience"
| (One intermediate revision by the same user not shown) | |||
| Line 6: | Line 6: | ||
===Applications of BBa_K1680009 by Grenoble 2019=== | ===Applications of BBa_K1680009 by Grenoble 2019=== | ||
| − | This is from the Grenoble 2019 team. For the NanoDrop project, the main problematic about the reporter gene was to choose between GFP and the Nanoluciferase reporter gene.We used Nanoluciferase following the Nano Glo Luciferase Assay System from Promega, which was offered to us within the scope of their sponsorship.The gene was received on a pNL1.1 vector. | + | This is from the Grenoble 2019 team. For the NanoDrop project, the main problematic about the reporter gene was to choose between GFP and the Nanoluciferase reporter gene.We used Nanoluciferase following the Nano Glo Luciferase Assay System from Promega, which was offered to us within the scope of their sponsorship.The gene was received on a pNL1.1 vector. |
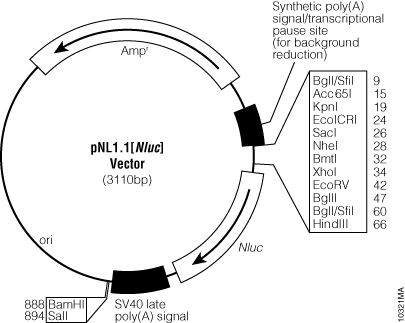
| − | This vector doesn’t contain a promoter. That is why a promoter has to be included in a plasmid to form a promoter-RBS-Nanoluciferase-terminator biobrick. For this, an inducible promoter is used in order to regulate the gene promoter with a signaling pathway. As seen in the BACTH document, an induction was generated thanks to the cAMP molecule. This is why a pLAC promoter is used : the lactose operon promoter. Indeed, it is regulated by cAMP and IPTG as it has been demonstrated with the BBa_J04450 part characterization. | + | [[File:Promega-vector-Nluc.jpeg|300px|thumb|right|Figure 1 : pNL1.1 vector from Promega.]] |
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | In this vector the nanoluciferase gene size is 513bp. | ||
| + | This vector doesn’t contain a promoter. That is why a promoter has to be included in a plasmid to form a promoter-RBS-Nanoluciferase-terminator biobrick. For this, an inducible promoter is used in order to regulate the gene promoter with a signaling pathway. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[File:Promoter-nanoluciferase.png|300px|thumb|left|Figure 2: Structure of the complete Lac operon and effect of cAMP on CAP promoter. To have more explanations, please refer you to the characterization document. ]] | ||
| + | |||
| + | |||
| + | As seen in the BACTH document, an induction was generated thanks to the cAMP molecule. This is why a pLAC promoter is used : the lactose operon promoter. Indeed, it is regulated by cAMP and IPTG as it has been demonstrated with the BBa_J04450 part characterization. | ||
| + | This building was made thanks to restriction site addition by PCR. This worked was made in endogeneous cAMP deficient bacteria, which are called BTH101. See more details on our wiki (https://2019.igem.org/Team:Grenoble-Alpes) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
Latest revision as of 12:48, 8 October 2019
This experience page is provided so that any user may enter their experience using this part.
Please enter
how you used this part and how it worked out.
Applications of BBa_K1680009 by Grenoble 2019
This is from the Grenoble 2019 team. For the NanoDrop project, the main problematic about the reporter gene was to choose between GFP and the Nanoluciferase reporter gene.We used Nanoluciferase following the Nano Glo Luciferase Assay System from Promega, which was offered to us within the scope of their sponsorship.The gene was received on a pNL1.1 vector.
In this vector the nanoluciferase gene size is 513bp.
This vector doesn’t contain a promoter. That is why a promoter has to be included in a plasmid to form a promoter-RBS-Nanoluciferase-terminator biobrick. For this, an inducible promoter is used in order to regulate the gene promoter with a signaling pathway.
As seen in the BACTH document, an induction was generated thanks to the cAMP molecule. This is why a pLAC promoter is used : the lactose operon promoter. Indeed, it is regulated by cAMP and IPTG as it has been demonstrated with the BBa_J04450 part characterization.
This building was made thanks to restriction site addition by PCR. This worked was made in endogeneous cAMP deficient bacteria, which are called BTH101. See more details on our wiki (https://2019.igem.org/Team:Grenoble-Alpes)
Applications of BBa_K1680009
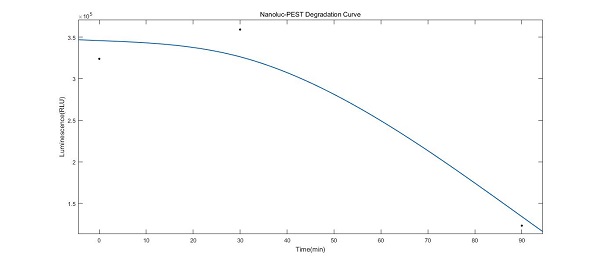
This is from Team Tianjin of iGEM2018. We added a tag after Nanoluc named PEST. PEST sequence can be found in variety of eukaryotic cells and the sequence we used was from the literature Engineered Luciferase Reporter from a Deep Sea Shrimp Utilizing a Novel Imidazopyrazinone Substrate. The covalent linkage of ubiquitin to lysine residues of substrate proteins is a common means used by eucaryotic cells to signal their degradation by the 26S proteasome, a multiprotease complex located in the cytoplasm and the nucleus. Decades ago it had been demonstrated that the PEST regions, enriched with Pro, Glu, Ser, and Thr, were identified to indeed control the ubiquitination of regulatory short-lived proteins.
Inspired by our time-course measurement for the KaiABC oscillatory system, we realized the importance of the sensitivity of reporter genes when used to characterize the variation in our engineering strains. With the knowledge that the instability of proteins is associated with the existence of the so-called PEST regions, which control the ubiquitination of regulatory short-lived proteins, we decided to optimize our NanoLuc with the potentially degrading sequence PSET to shorten its intracellular lifetime.
You can learn more details on this page.
User Reviews
UNIQ59b8f3210accec54-partinfo-00000000-QINU UNIQ59b8f3210accec54-partinfo-00000001-QINU