Difference between revisions of "Part:BBa K3182108"
| Line 81: | Line 81: | ||
In this study the calculated value of 22,22 µg per cm^2 sfGFP that could bind into the cellulose bandage. This bindning assay was not done at 4°C, which could lead to a less efficient binding capability of the protein. | In this study the calculated value of 22,22 µg per cm^2 sfGFP that could bind into the cellulose bandage. This bindning assay was not done at 4°C, which could lead to a less efficient binding capability of the protein. | ||
| − | |||
This was done by incubating 6 ml purified CBD-sfGFP solved in 650 ul 10x PBS with 0.2 g of a cellulosebandage with an area of 4.05 cm^2. The absorbance dropped 0.113 until the curve subside. Using this data and the molar absorbance of sfGFP at 481 nm, we could calculate the total amount of sfGFP that could bind into the bandage over time. The molar amount absorbed by the bandage was | This was done by incubating 6 ml purified CBD-sfGFP solved in 650 ul 10x PBS with 0.2 g of a cellulosebandage with an area of 4.05 cm^2. The absorbance dropped 0.113 until the curve subside. Using this data and the molar absorbance of sfGFP at 481 nm, we could calculate the total amount of sfGFP that could bind into the bandage over time. The molar amount absorbed by the bandage was | ||
Revision as of 12:51, 28 September 2019
Contents
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21INCOMPATIBLE WITH RFC[21]Illegal BamHI site found at 580
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 598
Introduction
pT7-CBDcipA-sfGFP
This part consists of a cellulose binding domain (CBD) from Clostridium thermocellum (C. thermocellum) cellulose scaffolding protein (CipA) and is a central part Clostridium thermocellum's cellusome. The CBD was fused to sfGFP in this part to easily track the binding capacities and to test our release mechanism. The CBD-sfGFP were fused using a flexible GS-linker (-GGGGSGGGGS-). A thrombin cleavage site (-LVPRGS-) was added to the end of the linker and its breakage will leave a glycine and serine attached to the N-terminal of the sfGFP fusion protein.
Assembly compabilities
An internal BamHI recognition sequence (RS) has been added to enable changeable fusion proteins. BamHI was chosen because its RS codes for glycine and serine, fitting it to the end of the thrombin site. It is also cost-effective enzyme and is unaffected by methylated DNA.
This part can be used to track purification, measure CBD binding ability and report cleavage at the thrombin site.
CBDcipA crystal structure
Important molecular faces
CBDcipA is composed of a nine-stranded beta sandwich with a jelly roll topology and binds a calcium ion. It further contains conserved residues exposed on the surface which map into two clear surfaces on each side of the molecule. One of faces mainly contains planar strips of aromatic and polar residues which may be the cellulose binding part. Further aspect are unknown and unique with this CBD such as the other conserved residues which are contained in a groove.
The choice of cellulose binding domain
iGEM Linköping 2019 choose CBDcipA due to many other iGEM teams exploring the possibilities of this domain. Our basic design was influenced by iGEM14 Imperial, iGEM15 Edinburgh and iGEM18 Ecuador. Purification and where to place the fusion protein (N- or C-terminal) was determined by studying the former projects. CBDcipA also originates from a thermophilic bacteria which further increases the domains applications.
Expression system
The part has a very strong expression with a T7-RNA-polymerase promotor (BBa_I719005) as well as a 5'-UTR (BBa_K1758100) region which has been shown to further increase expression in E. coli (BBa_K1758106), ([http://www.ncbi.nlm.nih.gov/pubmed/2676996 Olins et al. 1989]), ([http://www.ncbi.nlm.nih.gov/pubmed/23927491 Takahashi et al. 2013]). Both this part and the part were sfGFP was changed for AsPink (BBa_K3182000) showed great expression.

Usage and Biology
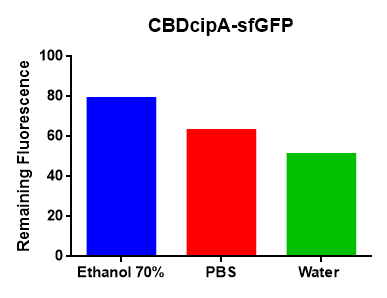

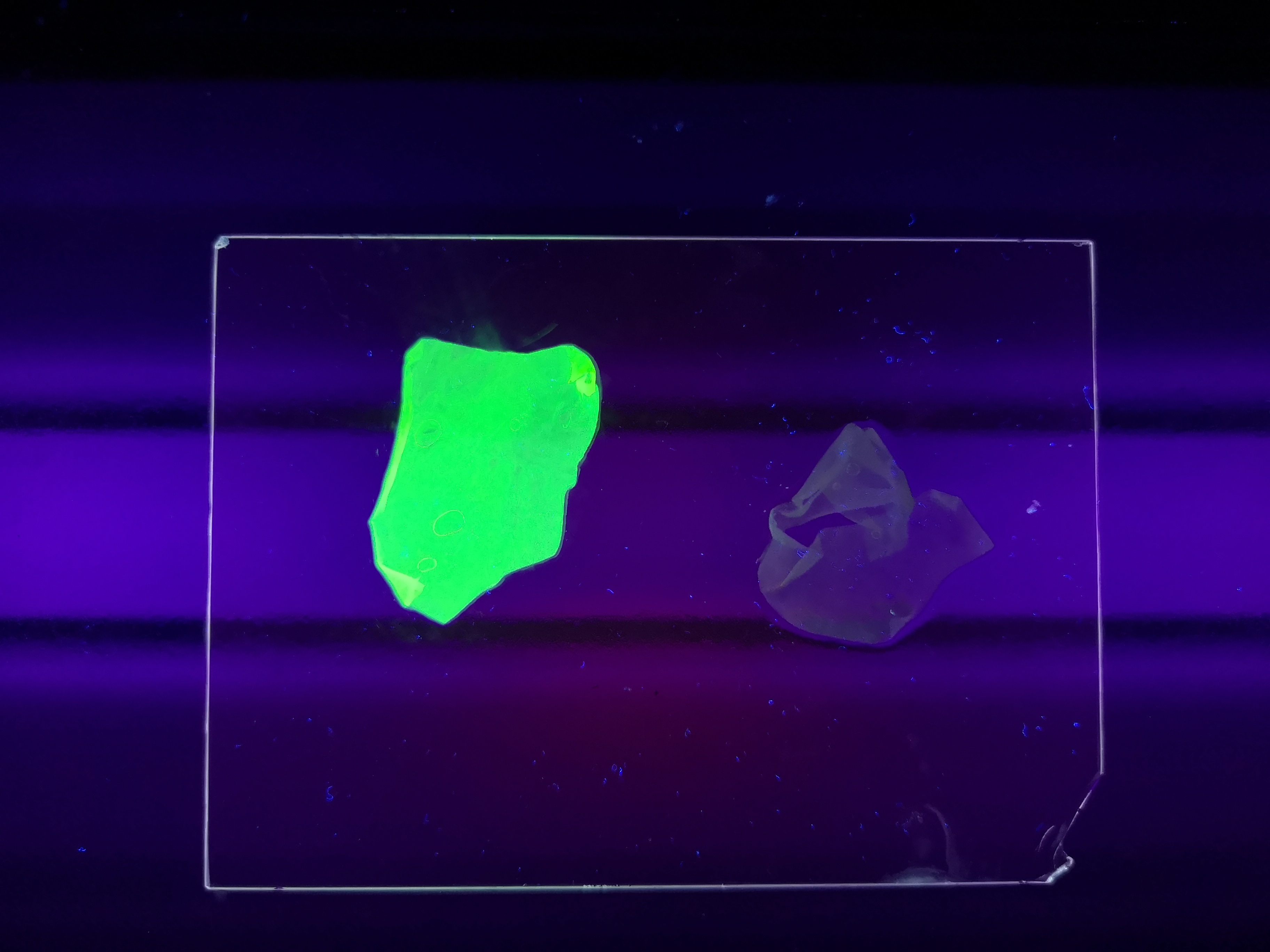 Figure Z Picture 1: Binding studies of the CBDcipA-sfGFP bound to bacterial cellulose. Washed three times with either 70 % ethanol, PBS or deionized water. Picture 2: Induced culture after 16 hours. E. coli BL21 (DE3) cells were grown in prescence of 25 ug/mL chlorampenicol until an OD600 of 0.8 at 37 degrees Celsius, and later induced with 0.5 mM IPTG. The induced culture were then incubated in 16 degrees Celsius for 16 hours. Picture 3: Left: CBDcipA-sfGFP bound to bacterial cellulose in form of a thin film, right: bacterial cellulose reference. Binding of CBDcipA-sfGFP was done the same way as the pictures below.
Figure Z Picture 1: Binding studies of the CBDcipA-sfGFP bound to bacterial cellulose. Washed three times with either 70 % ethanol, PBS or deionized water. Picture 2: Induced culture after 16 hours. E. coli BL21 (DE3) cells were grown in prescence of 25 ug/mL chlorampenicol until an OD600 of 0.8 at 37 degrees Celsius, and later induced with 0.5 mM IPTG. The induced culture were then incubated in 16 degrees Celsius for 16 hours. Picture 3: Left: CBDcipA-sfGFP bound to bacterial cellulose in form of a thin film, right: bacterial cellulose reference. Binding of CBDcipA-sfGFP was done the same way as the pictures below.
Figure A Picture 1: Lysate containing CBDcipA-sfGFP with bacterial cellulose before incubation. Picture 2: Lysate (CBDcipA-sfGFP) bound to bacterial cellulose after incubation in room temperature for 30 minutes on an end-to-end rotator. Picture 3: Bacterial cellulose after incubation with 70 % ethanol in room temperature for 30 minutes on an end-to-end rotator. All pictures were taken on a 302 nm UV-table for better visualization of the result.
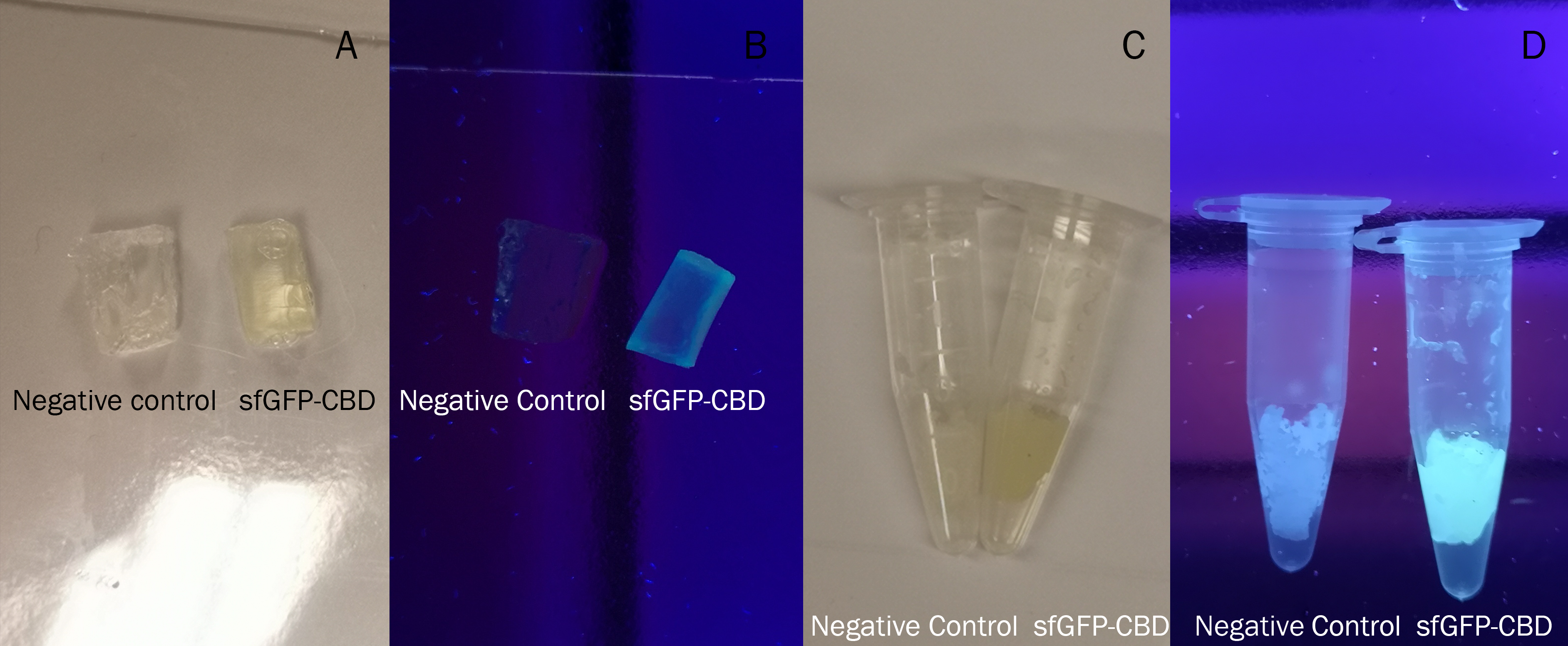
All of the photos above utilised the same BL21 (DE3) Gold CBD-sfGFP lysate and was incubated the same amount of time, 30min in room temperature. Both the CBD-sfGFP bound agarose and agarose that was not incubated CBD-sfGFP was washed in 70% ethanol once.
Purification of CBD-sfGFP
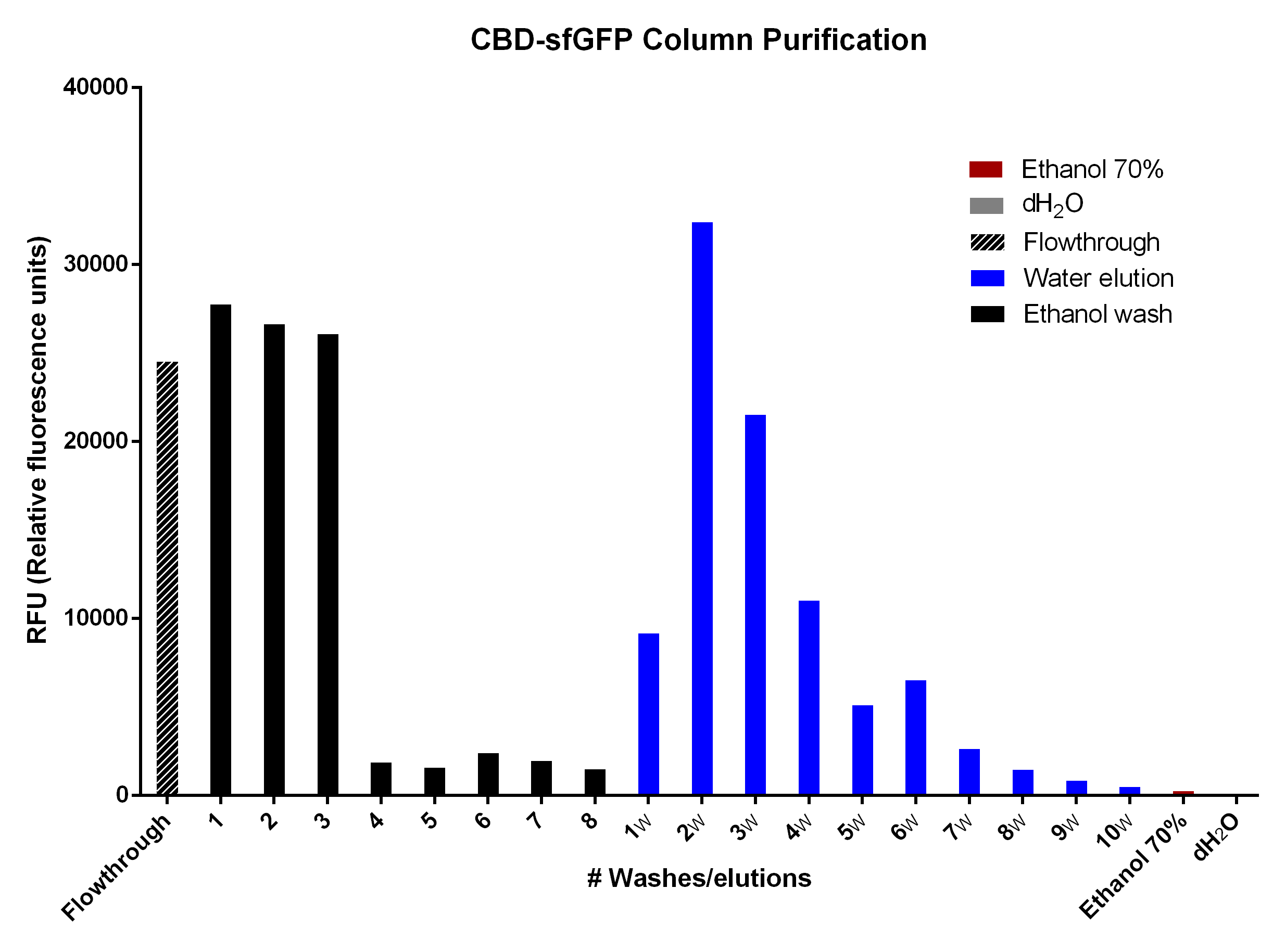
CBD-sfGFP was purified with cellulose fibres medium length (Whatman, #CF11) in a column. The first striped graph is the flowthrough after adding BL21 (De3) Gold lysate. The different washes of 70% ethanol was to remove unspecifically bound CBD-sfGFP, leading to the bound CBD-sfGFP remaining. This
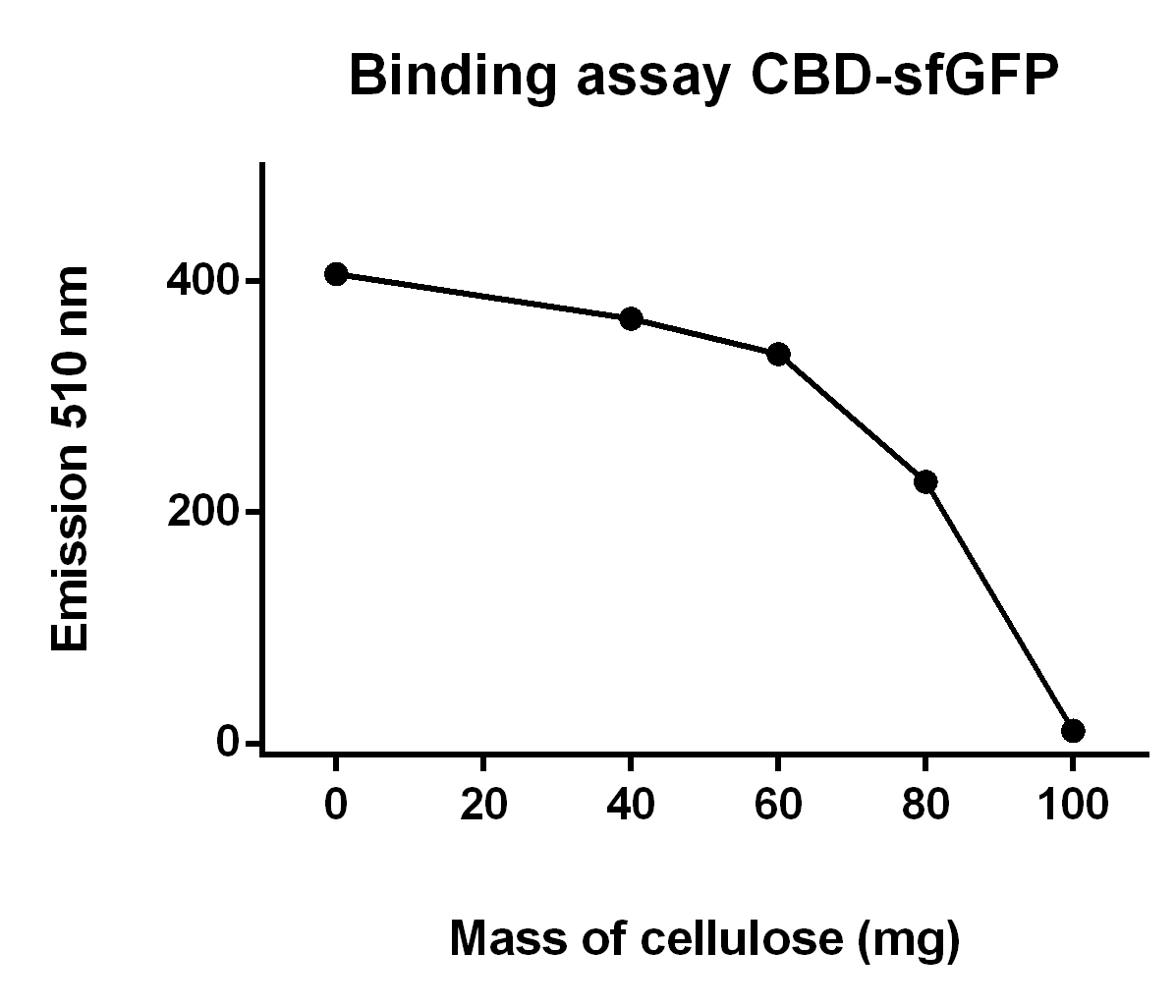
Binding assay of CBD-sfGFP
The illustration to the left, see figure X, shows that increasing amount of cellulose CF11 fibers increases the binding of CBD-sfGFP.
To make the binding assay Eppendorf tubes with varying mass of cellulose betweeb 0.00 - 1.00 g was mixed with a constant volume of CBD-sfGFP lysate (700 uL) and vortexed. The tubes was then attached to a end-to-end rotator for 30 minutes, thereafter they were placed in a tube holder until the cellulose powder settled in the bottom and the supernatant containing lysate was clear of cellulose. After that 100 µL from each of the supernatants was applied on a plate reader which measured the fluorescence of CBD-sfGFP at xx nm.
Total binding ability of CBD.sfGFP to cellulose bandage
The figure to the left resembles the binding capability of CBD-sfGFP to the cellulose bandage over time, measured by absorbance spectrometry at the 492.5 nm.
According to studies of sfGFP the absorption maximum is 485 nm, but in this study CBD-sfGFP had the absorption maximum at 492.5 nm. This leads to a change of absorption coefficient value (epsilon), which could not be found so instead a value of epsilon at 485 nm were used for calculating the binding capability of the protein. At 492.5 nm around ~93% of the electrons is excited which leads to an approximate value of the amount that could bind in to the cellulose surface per square cm.
In this study the calculated value of 22,22 µg per cm^2 sfGFP that could bind into the cellulose bandage. This bindning assay was not done at 4°C, which could lead to a less efficient binding capability of the protein.
This was done by incubating 6 ml purified CBD-sfGFP solved in 650 ul 10x PBS with 0.2 g of a cellulosebandage with an area of 4.05 cm^2. The absorbance dropped 0.113 until the curve subside. Using this data and the molar absorbance of sfGFP at 481 nm, we could calculate the total amount of sfGFP that could bind into the bandage over time. The molar amount absorbed by the bandage was
Reporter of successful cleavage and release from the cellulose binding domain
Spectrophotometrically analysis of thrombin cleavage
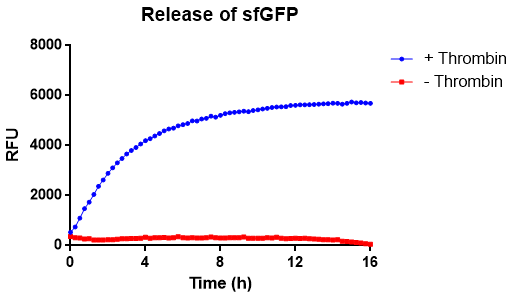
In figure X the release of sfGFP from our bacterial cellulose bandage can be seen over time. The cellulose-CBD-sfGFP were attached to the side of wells of a 96-well plate and 200 uL 1X thrombin cleavage buffer (20 mM Tris-HCl, 150 mM NaCl and 2.5 mM CaCl2) were added. To the wells with cellulose-CBDcipA-sfGFP and buffer, 0.03 units of human thrombin were added and fluorescence (ex. 485 nm, em. 510 nm) were measured from the bottom and up (center of the well) for 16 hours. The temperature was set to 37 degrees Celsius. In blue, successful release of sfGFP from the CBD can be seen. In red, the control experiment can be seen, where no thrombin was added.
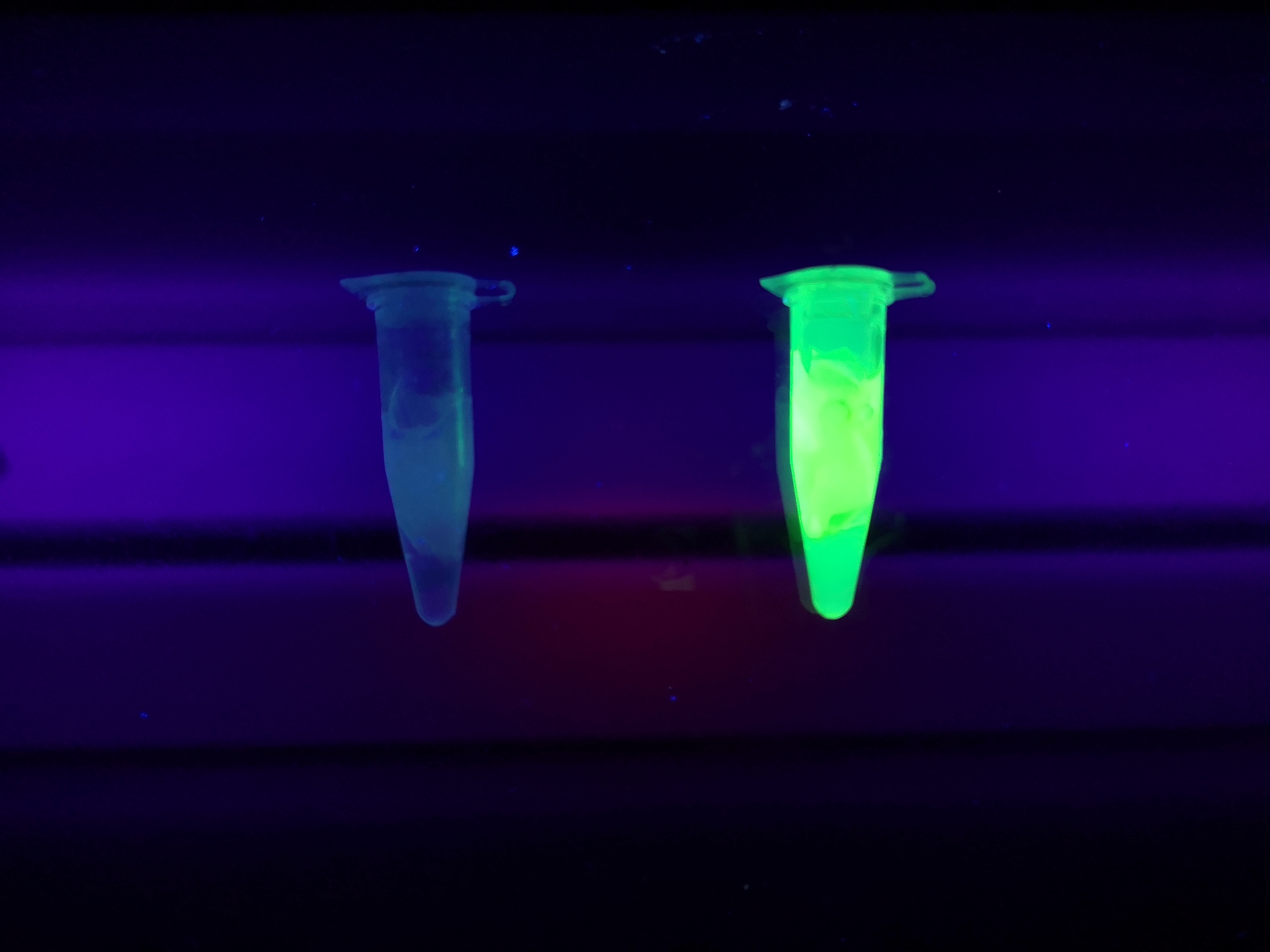
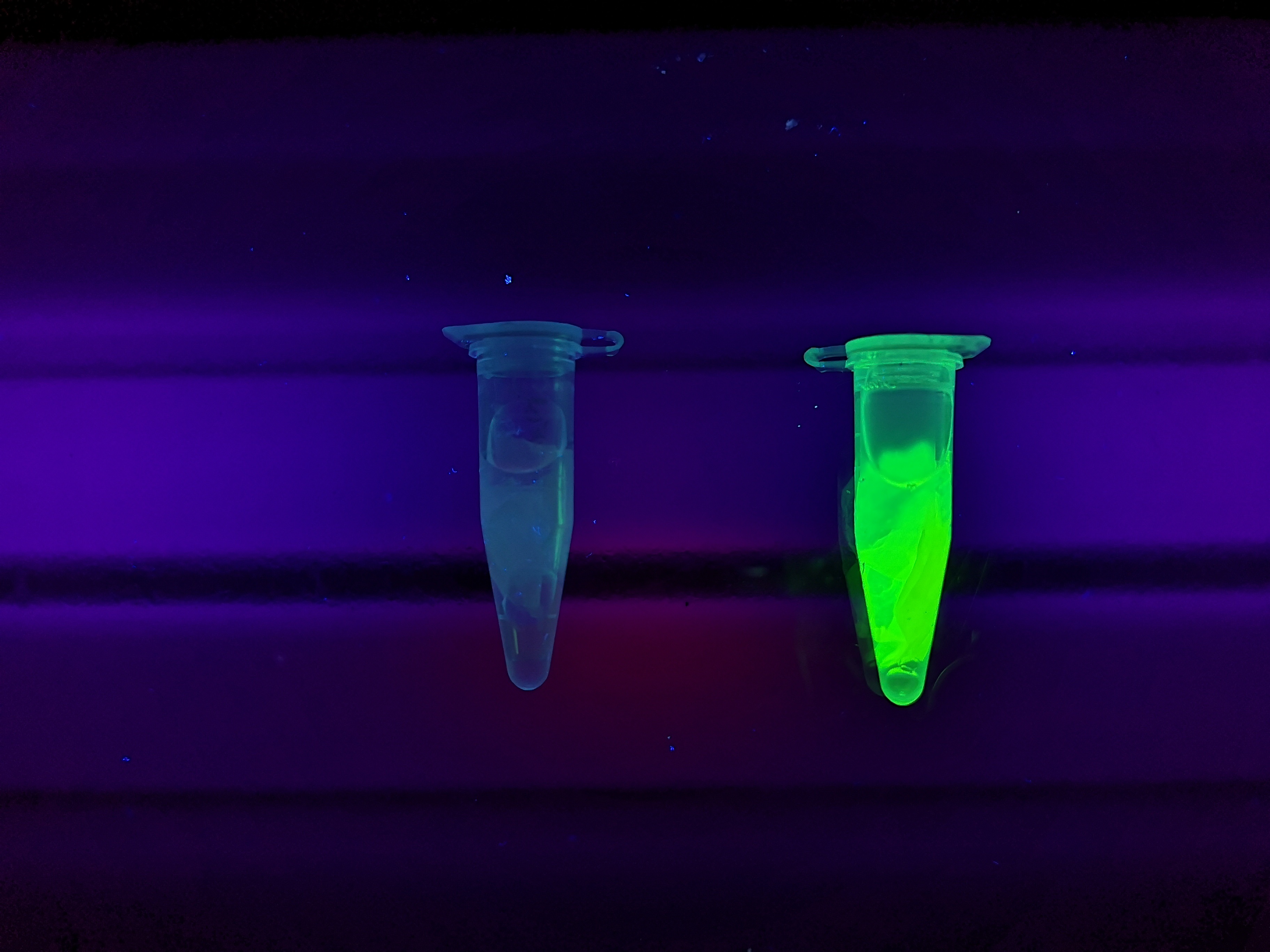
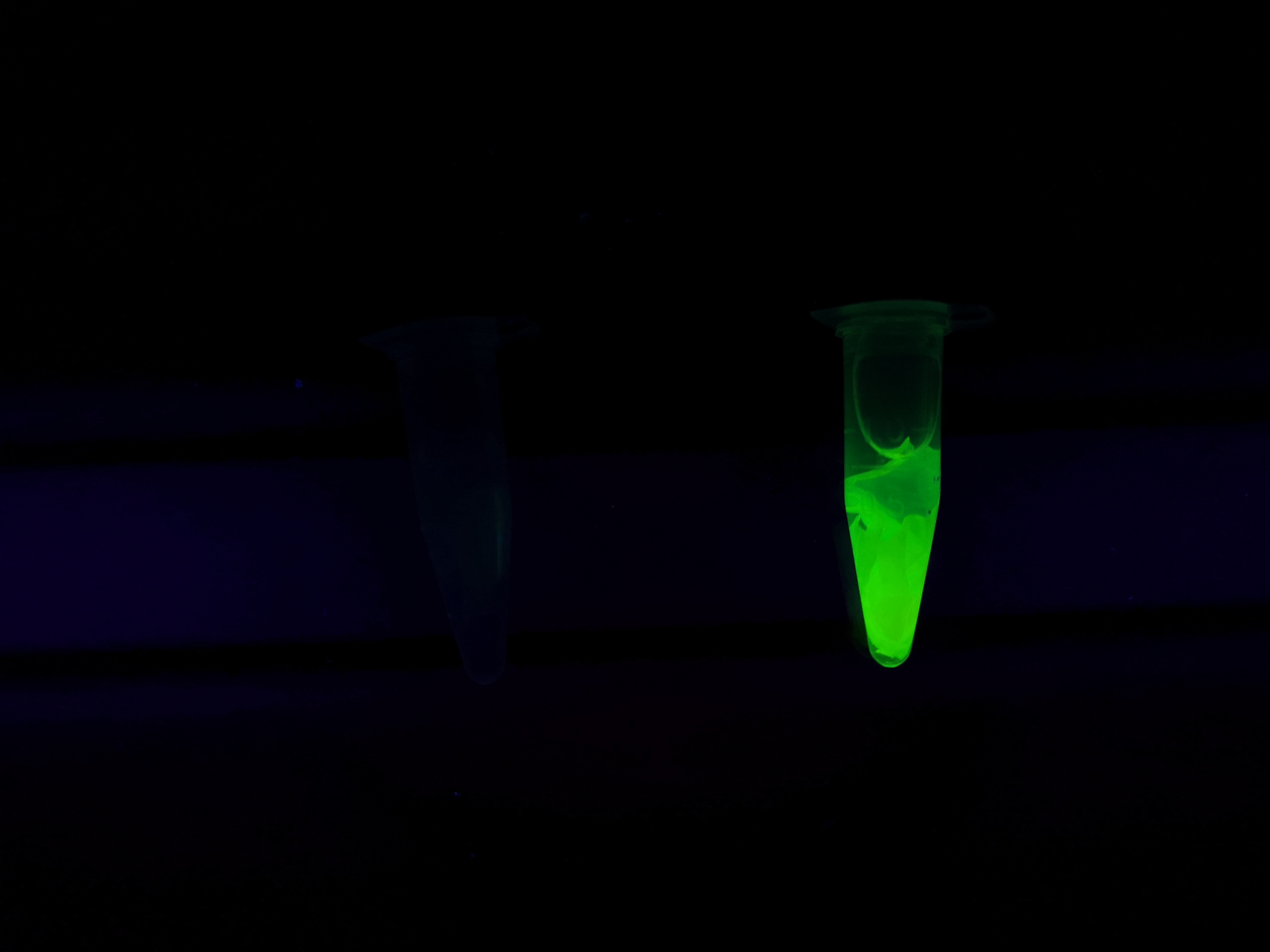
Visual experiment of thrombin cleavage
To the left a visual experiment with this part can be seen. After unbound protein had been removed the cellulose was washed three times with 70 % ethanol. To test the activity, 200 uL thrombin cleavage buffer (20 mM Tris-HCl, 150 mM NaCl and 2.5 mM CaCl2) were added along side 0.03 units of human thrombin to the bacterial cellulose. To the right in the figure, the successful cleavage of CBDcipA-sfGFP can be seen. The cellulose is to the left of the tube where free (cleaved at the thrombin site) sfGFP can be seen. To the left, the control sample can be seen, where no sfGFP can be seen in the supernatant. The picture is taken on a 302 nm UV-table to better visualize the results.
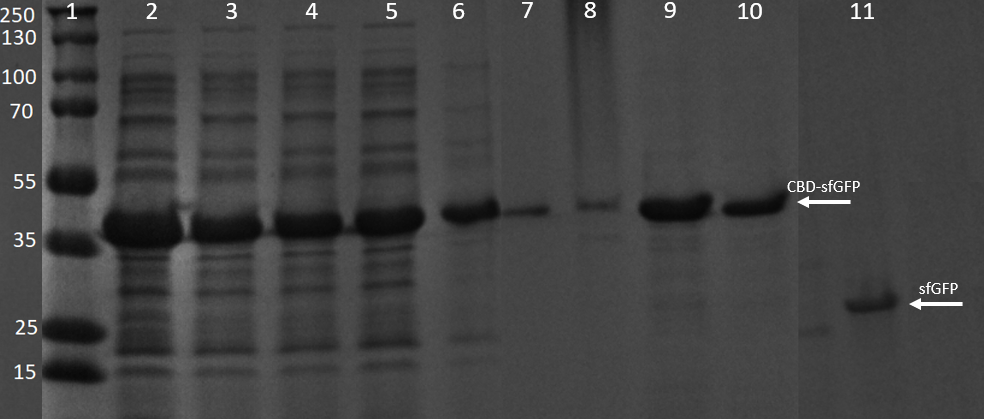
SDS-PAGE analysis of cleavage and expression
E. coli BL21 (DE3) cells were grown in prescence of 25 ug/mL chlorampenicol until an OD600 of 0.8 at 37 degrees Celsius, and later induced with 0.5 mM IPTG. The induced culture were then incubated in 16 degrees Celsius for 16 hours. The bacteria was then lysed with sonication at 30 % for 6 minutes. Most of this part could be found in the soluble fraction. The lysate (1 mL) was then incubated with cellulose (CF11) for 30 minutes in room temperature. Four washes with 70 % ethanol was then conducted all with a volume of 1 mL. Elution of CBDcipA-sfGFP was done with 1 mL fractions of dH2O. One replicate was instead cleaved with thrombin instead. Thrombin cleavage buffer (20 mM Tris-HCl, 150 mM NaCl and 2.5 mM CaCl2) was added to the Eppendorf tube with the cellulose at a volume of 500 uL, and 0.03 units was then added to the solution. The cleavage was done in room temperature over 16 hours, with inversion of the tube.