Difference between revisions of "Loop Assembly"
m |
m |
||
| (42 intermediate revisions by 2 users not shown) | |||
| Line 1: | Line 1: | ||
| − | + | ==Request For Comments== | |
| − | As we prepare for iGEM 2019, | + | As we prepare for iGEM 2019, we have been trying to decide how to support Type IIS assembly methods for the Registry and the 2019 iGEM Competition. Since [http://2016.igem.org/Resources/Plant_Synthetic_Biology/PhytoBricks 2016], iGEM has allowed teams to submit parts in the PhytoBricks (Type IIS Golden Gate) standard, but we have not fully supported any Type IIS standard. |
Our current proposal is as follows: | Our current proposal is as follows: | ||
| − | <blockquote>iGEM fully adopts the [https://www.biorxiv.org/content/early/2018/01/18/247593 Loop Assembly standard] with Level 0 (basic) parts adhering to the MoClo and/or [https://nph.onlinelibrary.wiley.com/doi/full/10.1111/nph.13532 PhytoBricks] Golden Gate assembly standards. | + | <blockquote>iGEM fully adopts the [https://www.biorxiv.org/content/early/2018/01/18/247593 Loop Assembly standard] with Level 0 (basic) parts adhering to the [https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0016765 MoClo] and/or [https://nph.onlinelibrary.wiley.com/doi/full/10.1111/nph.13532 PhytoBricks] Golden Gate assembly standards. |
Adoption of Loop would include: | Adoption of Loop would include: | ||
| Line 17: | Line 17: | ||
| − | Before we finalize the plans for adopting Loop, I would like to collect input from our community. If you have any comments or thoughts on iGEM adopting and supporting Loop (including the MoClo and/or PhytoBricks assembly standards), please send an email to | + | Before we finalize the plans for adopting Loop, I would like to collect input from our community. If you have any comments or thoughts on iGEM adopting and supporting Loop (including the MoClo and/or PhytoBricks assembly standards), please send an email to '''registry (at) igem (dot) org''' by Tuesday, December 4th 2018. |
| − | + | ||
For more information please read below and visit the linked resources. | For more information please read below and visit the linked resources. | ||
| + | <div style="text-align: right">''Vinoo Selvarajah, Director of the Registry''</div> | ||
| − | |||
| − | |||
| − | + | ==Why Loop?== | |
| − | The | + | The BioBrick RFC [10] Standard, developed by Tom Knight, has been incredibly successful for well over a decade. With BioBrick parts and 3A Assembly, a synthetic biologist could reliably assemble their parts together without having to do an experiment each time. It just worked, so engineers could focus on their designs. |
| − | + | The standard is not without its issues and limitations though, and there have been new assembly methods since then that addressed these. The value of BioBricks parts and assembly still remains; it's easy, reliable, widespread, and parts are completely interchangeable. | |
| + | That said, Type IIS assembly methods address some of the limitations of BioBricks while maintaining similar value. Parts are not completely interchangeable (nor is the standard idempotent), but there is flexibility when new samples of parts are designed to take advantage of user-specified fusion sites/positioning. Most importantly, multiple parts can be assembled easily and reliably in a single reaction, and this can be scaled further to create more complex genetic circuits very quickly. As the problems iGEM teams tackle become more complex, an assembly strategy that would allow they to design and rapidly construct multiple genetic circuits quickly is certainly attractive. | ||
| − | |||
| + | Currently, there are several Type IIS assembly standards in use, many of these were designed for specific purposes. iGEM believes strongly in standards, and as we approach Type IIS assembly methods we want to support one that we believe will be useful, reliable, long-lasting and widespread for the synthetic biology community. | ||
| − | |||
| − | |||
| − | |||
===Adoption=== | ===Adoption=== | ||
| − | We know several groups, including academic | + | We know several groups, including academic, research labs and industry, that are already using Loop Assembly. We want to ensure that the assembly method we support is widely used, tested, and has flexibility for different applications and groups. |
| + | |||
| + | Tom Knight occasionally jokes that "the nice thing about standards is that there are so many to choose from." In reality, sharing only works when a community adopts the same standard. There is a network effect that makes the collection much more valuable than the pieces. For this reason, we hope to adopt an existing approach instead of developing a new one. | ||
Loop Assembly is compatible with a variety of Golden Gate standards for Level 0 parts, most importantly MoClo and PhytoBricks. Both of these standards are widely used, and MoClo's fusion sites completely overlap with PhytoBricks. | Loop Assembly is compatible with a variety of Golden Gate standards for Level 0 parts, most importantly MoClo and PhytoBricks. Both of these standards are widely used, and MoClo's fusion sites completely overlap with PhytoBricks. | ||
| + | |||
| + | |||
| Line 50: | Line 50: | ||
Loop uses two Type IIS sites for assembly: BsaI (6bp recognition site) and SapI (a 7bp recognition site). Parts designed to be used in Loop only need to be free of these two restriction sites, meaning theoretically part standardization should be easier than BioBrick RFC10 (4 illegal restriction sites). | Loop uses two Type IIS sites for assembly: BsaI (6bp recognition site) and SapI (a 7bp recognition site). Parts designed to be used in Loop only need to be free of these two restriction sites, meaning theoretically part standardization should be easier than BioBrick RFC10 (4 illegal restriction sites). | ||
| − | Like other | + | Like other Type IIS assembly methods, Loop is structured to allow for increasing complexity of assemblies (Level 1s and 2s). However, the recursive design in Loop allows for the same two cloning vectors to be used to easily create larger and more complex circuits (Level 3 and 4s). |
| + | The Loop Assembly vectors are also under the OpenMTA license for unrestricted sharing. iGEM can distribute and modify these vectors as needed, and so can iGEM teams/groups. | ||
| − | ===OpenMTA=== | + | |
| − | The Loop Assembly vectors outlined in the paper are under the OpenMTA license for unrestricted sharing. iGEM | + | |
| + | |||
| + | ==Background on Type IIS Assembly== | ||
| + | Type IIS assembly methods use Type IIS restriction enzymes, which cleave DNA outside their recognition site. First introduced in Golden Gate cloning ([https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0003647 Engler et al. 2008]), these assembly methods standardize the cleavage sites of the Type IIS enzymes as "fusion sites." Basic parts (Promoter, RBS, CDS, etc) are flanked by defined fusion sites which are dictated by the part type and/or desired position for assembly. When part samples are cut with a Type IIS enzyme they will produce compatible overhangs at the fusion sites which will allow the parts to be ligated in the desired order. This means that a single one-pot assembly reaction can include multiple basic parts (Promoter, RBS, CDS, Terminator) and result in a complete transcriptional unit (composite part). | ||
| + | |||
| + | See the diagram below for an example of a Type IIS assembly reaction. | ||
| + | |||
| + | [[File:Assembly_-_GoldenGate_Example.jpeg|thumb|center|Example of Type IIS enzyme (BsaI) cleaving multiple parts for assembly (Fig 1 from Patron et al. 2015)]] | ||
| + | |||
| + | |||
| + | ===Level 0 (basic parts)=== | ||
| + | The MoClo assembly standard ([https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0016765 Weber et al. 2011]) built upon Golden Gate and defined a standard by which parts could be assembled in increasing complexity: Level 0 as basic parts, Level 1 as transcriptional units (composite parts), and Level 2 as multi-transcriptional unit assemblies. | ||
| + | |||
| + | The PhytoBricks assembly standard ([https://nph.onlinelibrary.wiley.com/doi/full/10.1111/nph.13532 Patron et al. 2015]) built upon Golden Gate and MoClo to develop an assembly standard for the plant synthetic biology community. This Type IIS standard introduced additional fusion sites and a new framework to accommodate the needs of synthetic biology for eukaryotic organisms. The standard is widely used in the plant synthetic biology community and has been an accepted standard for submissions for iGEM teams since 2016. | ||
| + | |||
| + | <center><gallery widths=250px> | ||
| + | File:Assembly_-_MoClo_Diagram.png|MoClo parts and fusion sites (Figure 1 from Weber et al. 2011) | ||
| + | File:Assembly_-_PhytoBricks_Standard.jpeg|PhytoBrick parts and fusion sites (Figure 3 from Patron et al. 2015) | ||
| + | </gallery></center> | ||
| + | |||
| + | For 2019, we plan to accept Level 0 (Promoters, RBS, etc) parts that adhere to the PhytoBricks and MoClo standards. We have compared the fusion sites used in both standards with the work done by NEB and Ginkgo ([https://pubs.acs.org/doi/10.1021/acssynbio.8b00333 Potapov et al. 2018]) on fusion site fidelity. All fusion sites except one used in PhytoBricks show high fidelity ligation when used as a set. | ||
| + | |||
| + | |||
| + | ===Loop Assembly=== | ||
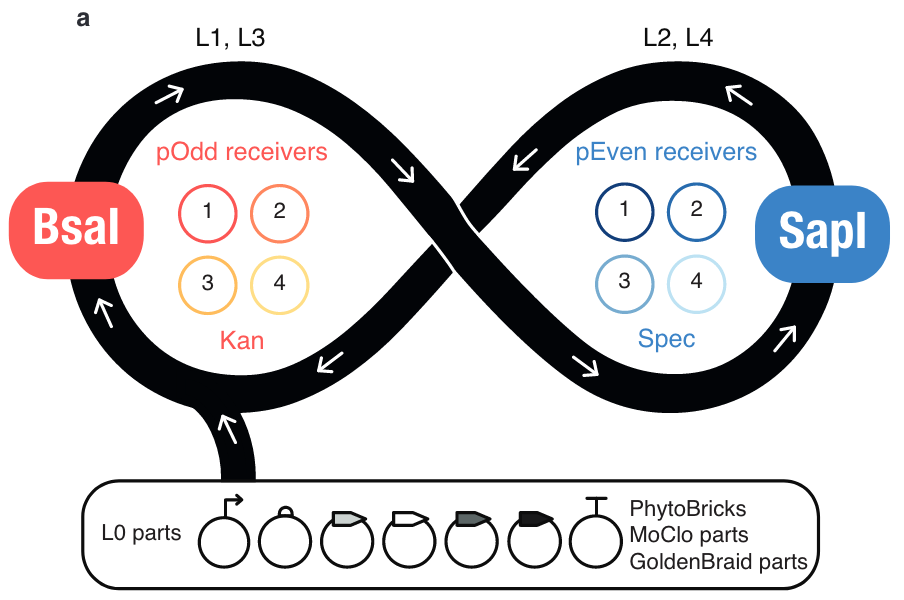
| + | |||
| + | [[File:Assembly_-_Loop_General_Design.png|thumb|center|(Fig 1 from Pollak et al. 2018]] | ||
| + | |||
| + | Loop Assembly ([https://www.biorxiv.org/content/early/2018/01/18/247593 Pollak et al. 2018])is completely compatible with the basic parts (Level 0s) used in the PhytoBricks and MoClo standard. The Loop standard addresses the needs of creating more complex and larger assemblies (Level 2+) using a recursive approach to assembly. Using two vectors with alternating Type IIS restriction sites, samples of MoClo/PhytoBrick Level 0 parts can be assembled into 1 transcriptional unit, then up to 4, 16, or more. | ||
| + | |||
| + | |||
| + | For more information on Loop Assembly, developed at the [http://www.haseloff-lab.org/ Haseloff Lab], please read through the [https://www.biorxiv.org/content/early/2018/01/15/247593 manuscript on biorxiv]. | ||
| + | |||
| + | |||
| + | |||
| + | <!--===OpenMTA=== | ||
| + | The Loop Assembly vectors outlined in the paper are under the OpenMTA license for unrestricted sharing. iGEM can distribute and modify these vectors as needed, and so can iGEM teams. For more information on the OpenMTA license, developed by [https://www.openplant.org/home2/ OpenPlant] and [https://biobricks.org/ The BioBricks Foundation], please read--> | ||
| + | |||
| + | __NOTOC__ | ||
Latest revision as of 22:33, 30 November 2018
Request For Comments
As we prepare for iGEM 2019, we have been trying to decide how to support Type IIS assembly methods for the Registry and the 2019 iGEM Competition. Since [http://2016.igem.org/Resources/Plant_Synthetic_Biology/PhytoBricks 2016], iGEM has allowed teams to submit parts in the PhytoBricks (Type IIS Golden Gate) standard, but we have not fully supported any Type IIS standard.
Our current proposal is as follows:
iGEM fully adopts the Loop Assembly standard with Level 0 (basic) parts adhering to the MoClo and/or PhytoBricks Golden Gate assembly standards.Adoption of Loop would include:
- giving teams full credit for submitting samples in this standard
- updating the Registry software to support the standard
- creating documentation for our users
- distributing a collection of basic parts and cloning vectors in the 2019 Distribution Kit.
iGEM would, of course, continue to support and send out BioBrick parts for the foreseeable future.
Before we finalize the plans for adopting Loop, I would like to collect input from our community. If you have any comments or thoughts on iGEM adopting and supporting Loop (including the MoClo and/or PhytoBricks assembly standards), please send an email to registry (at) igem (dot) org by Tuesday, December 4th 2018.
For more information please read below and visit the linked resources.
Why Loop?
The BioBrick RFC [10] Standard, developed by Tom Knight, has been incredibly successful for well over a decade. With BioBrick parts and 3A Assembly, a synthetic biologist could reliably assemble their parts together without having to do an experiment each time. It just worked, so engineers could focus on their designs.
The standard is not without its issues and limitations though, and there have been new assembly methods since then that addressed these. The value of BioBricks parts and assembly still remains; it's easy, reliable, widespread, and parts are completely interchangeable.
That said, Type IIS assembly methods address some of the limitations of BioBricks while maintaining similar value. Parts are not completely interchangeable (nor is the standard idempotent), but there is flexibility when new samples of parts are designed to take advantage of user-specified fusion sites/positioning. Most importantly, multiple parts can be assembled easily and reliably in a single reaction, and this can be scaled further to create more complex genetic circuits very quickly. As the problems iGEM teams tackle become more complex, an assembly strategy that would allow they to design and rapidly construct multiple genetic circuits quickly is certainly attractive.
Currently, there are several Type IIS assembly standards in use, many of these were designed for specific purposes. iGEM believes strongly in standards, and as we approach Type IIS assembly methods we want to support one that we believe will be useful, reliable, long-lasting and widespread for the synthetic biology community.
Adoption
We know several groups, including academic, research labs and industry, that are already using Loop Assembly. We want to ensure that the assembly method we support is widely used, tested, and has flexibility for different applications and groups.
Tom Knight occasionally jokes that "the nice thing about standards is that there are so many to choose from." In reality, sharing only works when a community adopts the same standard. There is a network effect that makes the collection much more valuable than the pieces. For this reason, we hope to adopt an existing approach instead of developing a new one.
Loop Assembly is compatible with a variety of Golden Gate standards for Level 0 parts, most importantly MoClo and PhytoBricks. Both of these standards are widely used, and MoClo's fusion sites completely overlap with PhytoBricks.
Flexibility and Complexity
Loop uses two Type IIS sites for assembly: BsaI (6bp recognition site) and SapI (a 7bp recognition site). Parts designed to be used in Loop only need to be free of these two restriction sites, meaning theoretically part standardization should be easier than BioBrick RFC10 (4 illegal restriction sites).
Like other Type IIS assembly methods, Loop is structured to allow for increasing complexity of assemblies (Level 1s and 2s). However, the recursive design in Loop allows for the same two cloning vectors to be used to easily create larger and more complex circuits (Level 3 and 4s).
The Loop Assembly vectors are also under the OpenMTA license for unrestricted sharing. iGEM can distribute and modify these vectors as needed, and so can iGEM teams/groups.
Background on Type IIS Assembly
Type IIS assembly methods use Type IIS restriction enzymes, which cleave DNA outside their recognition site. First introduced in Golden Gate cloning (Engler et al. 2008), these assembly methods standardize the cleavage sites of the Type IIS enzymes as "fusion sites." Basic parts (Promoter, RBS, CDS, etc) are flanked by defined fusion sites which are dictated by the part type and/or desired position for assembly. When part samples are cut with a Type IIS enzyme they will produce compatible overhangs at the fusion sites which will allow the parts to be ligated in the desired order. This means that a single one-pot assembly reaction can include multiple basic parts (Promoter, RBS, CDS, Terminator) and result in a complete transcriptional unit (composite part).
See the diagram below for an example of a Type IIS assembly reaction.
Level 0 (basic parts)
The MoClo assembly standard (Weber et al. 2011) built upon Golden Gate and defined a standard by which parts could be assembled in increasing complexity: Level 0 as basic parts, Level 1 as transcriptional units (composite parts), and Level 2 as multi-transcriptional unit assemblies.
The PhytoBricks assembly standard (Patron et al. 2015) built upon Golden Gate and MoClo to develop an assembly standard for the plant synthetic biology community. This Type IIS standard introduced additional fusion sites and a new framework to accommodate the needs of synthetic biology for eukaryotic organisms. The standard is widely used in the plant synthetic biology community and has been an accepted standard for submissions for iGEM teams since 2016.
For 2019, we plan to accept Level 0 (Promoters, RBS, etc) parts that adhere to the PhytoBricks and MoClo standards. We have compared the fusion sites used in both standards with the work done by NEB and Ginkgo (Potapov et al. 2018) on fusion site fidelity. All fusion sites except one used in PhytoBricks show high fidelity ligation when used as a set.
Loop Assembly
Loop Assembly (Pollak et al. 2018)is completely compatible with the basic parts (Level 0s) used in the PhytoBricks and MoClo standard. The Loop standard addresses the needs of creating more complex and larger assemblies (Level 2+) using a recursive approach to assembly. Using two vectors with alternating Type IIS restriction sites, samples of MoClo/PhytoBrick Level 0 parts can be assembled into 1 transcriptional unit, then up to 4, 16, or more.
For more information on Loop Assembly, developed at the [http://www.haseloff-lab.org/ Haseloff Lab], please read through the manuscript on biorxiv.