Difference between revisions of "Part:BBa K2886004"
| (6 intermediate revisions by the same user not shown) | |||
| Line 23: | Line 23: | ||
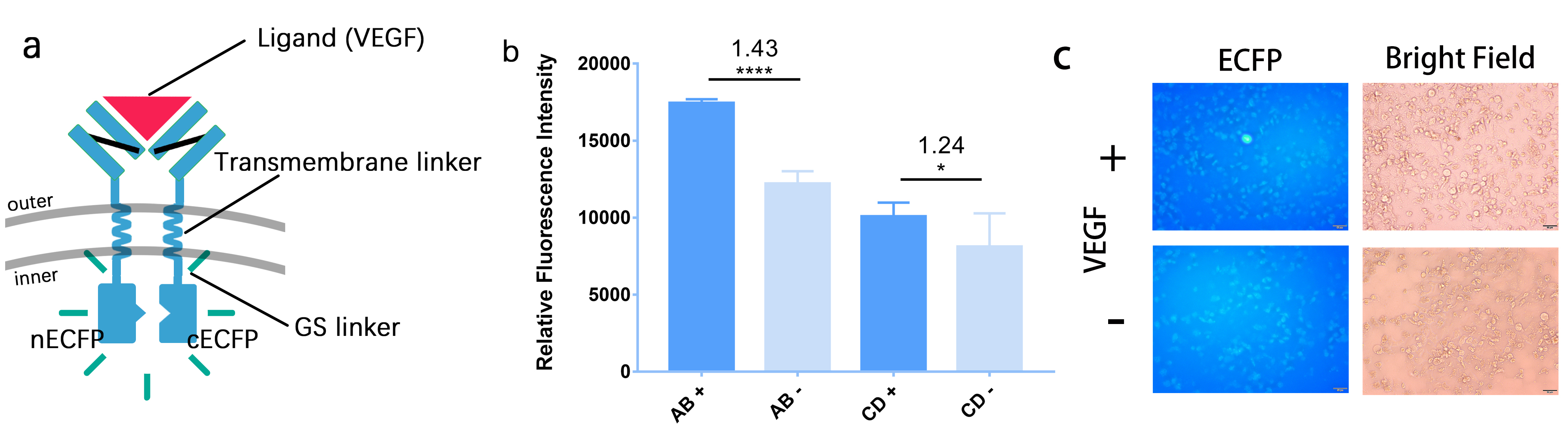
Moreover, to further demonstrate the reliability of our BiFC system, we also do some similar things in 293T cells. The difference is that we add outer-membrane and transmembrane domain to it as well as substitute the leucine zipper with GS linker. We transfect the cells with these plasmids and then induce them with the ligand VEGF. When we observe the cells under fluorescence microscope, we also see the fluorescence, which is showed in Figure 2. | Moreover, to further demonstrate the reliability of our BiFC system, we also do some similar things in 293T cells. The difference is that we add outer-membrane and transmembrane domain to it as well as substitute the leucine zipper with GS linker. We transfect the cells with these plasmids and then induce them with the ligand VEGF. When we observe the cells under fluorescence microscope, we also see the fluorescence, which is showed in Figure 2. | ||
| − | [[File: | + | [[File:Figure2_for_cECFP.jpeg|700px|thumb|left|'''Figure 2:''' The structure of BiFC in Hela cell and the results of tests. (a)We construct 4 plasmid, whose inner-membrane domains are A: 1 GS linker + cECFP, B: 1 GS linker + nECFP,C: 2 GS linkers + cECFP and D: 2 GS linkers + nECFP. (b)The fluorescence intensity of after adding VEGF (AB+) has a 1.43 folds compared with before (AB-) in the A+B group. And in C+D group, it’s 1.24 folds.(c)Relative fluorescence intensity at different cotransfection in A+B group. Relative fluorescence intensity is evaluated by grey level measurement of ImageJ. They all show apparent differences, so it’s safe to conclude that our BiFC system works.]] |
<br style="clear: both" /> | <br style="clear: both" /> | ||
| Line 32: | Line 32: | ||
===Improvement on a previous part=== | ===Improvement on a previous part=== | ||
In 2004, ECFP(engineered cyan fluorescent protein derived from A. victoria GFP) was introduced to iGEM by Antiquity in [[Part:BBa_E0020]]. Since then, it has had more than 80 uses. This year Fudan-CHINA improves it by taking it apart using BiFC (bimolecular fluorescence complementation) to tremendously expandthe usage of the original part [[Part:BBa_E0020]]. With improved parts, you can use them as labels for interaction of two target proteins and the location of two proteins in subcellular level. | In 2004, ECFP(engineered cyan fluorescent protein derived from A. victoria GFP) was introduced to iGEM by Antiquity in [[Part:BBa_E0020]]. Since then, it has had more than 80 uses. This year Fudan-CHINA improves it by taking it apart using BiFC (bimolecular fluorescence complementation) to tremendously expandthe usage of the original part [[Part:BBa_E0020]]. With improved parts, you can use them as labels for interaction of two target proteins and the location of two proteins in subcellular level. | ||
| + | |||
| + | For more detailed information, please click here[[http://2018.igem.org/Team:Fudan-CHINA/Improve]] to read our wiki. | ||
<!-- --> | <!-- --> | ||
Latest revision as of 20:34, 17 October 2018
C-terminal of ECFP(cECFP)
Usage and Biology
The cECFP was the C-terminal of ECFP. It could fluoresce when binding the N-terminal of ECFP(nECFP)
BBa_K2886004 together with Part:BBa_K2886005 are both improved from Part:BBa_E0020, which has 87 uses till 2017. And we designed them from BBa_E0020 with the technology BiFC (bimolecular fluorescence complementation).
BiFC is a new technology put forward in 2012 aiming to visualize the interaction of two proteins. If you connect these two parts with your two target proteins respectively, when two target proteins interact with each other, two parts of BiFC might well bind with each other and emit fluorescence for observation, which, in turn, proves the combination of your target proteins.
We split ECFP at the site A155 into two parts and get cECFP (coded by BBa_K2886004) and nECFP (coded by BBa_K2886005) are the C- and N-terminal of ECPF. If these two parts of BiFC bind with each other, and they will emit fluorescence just as complete ECFP.
To demonstrate that our BiFC system works, we carry out a series of experiment.
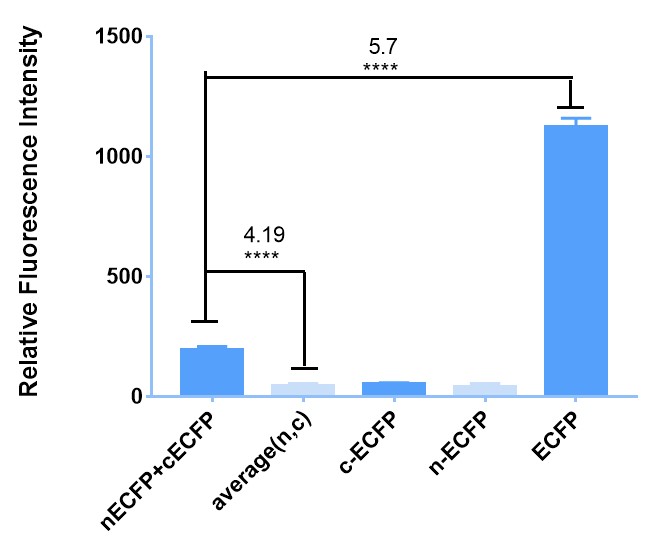
Firstly, to prove it preliminarily, we implement experiment in E. coli BL21. To begin with, we transfect E. coli with plasmids carrying BBa_K2886013 (leucine zipper + nECFP) and BBa_K2886012 (leucine zipper + cECFP) respectively. Because of the existence of leucine zipper, which can bind with each other due to electrostatic attraction automatically without out outer-membrane and transmembrane domain or ligand. Then, when they express enough cECFP and nECFP, we lyse the bacteria with ultrasound and mix them up to emit cECFP and nECFP to a same system. At last, we measure the fluorescence with plate reader compared with negative and positive controls. The Figure 1 shows that our attempt does succeed and it might be reliable for more complicated conditions.
Moreover, to further demonstrate the reliability of our BiFC system, we also do some similar things in 293T cells. The difference is that we add outer-membrane and transmembrane domain to it as well as substitute the leucine zipper with GS linker. We transfect the cells with these plasmids and then induce them with the ligand VEGF. When we observe the cells under fluorescence microscope, we also see the fluorescence, which is showed in Figure 2.
Through these two experiments, we fully demonstrate that our BiFC system works and prove the improvement of BBa_E0020 is a success. And BBa_K2886004 & BBa_K2886005 can used as labels for interaction of two target proteins and the location of two proteins in subcellular level, tremendously expanding the usage of the original part and equipping iGEMers another powerful tool for biological visualization.
Improvement on a previous part
In 2004, ECFP(engineered cyan fluorescent protein derived from A. victoria GFP) was introduced to iGEM by Antiquity in Part:BBa_E0020. Since then, it has had more than 80 uses. This year Fudan-CHINA improves it by taking it apart using BiFC (bimolecular fluorescence complementation) to tremendously expandthe usage of the original part Part:BBa_E0020. With improved parts, you can use them as labels for interaction of two target proteins and the location of two proteins in subcellular level.
For more detailed information, please click herehttp://2018.igem.org/Team:Fudan-CHINA/Improve to read our wiki.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000COMPATIBLE WITH RFC[1000]
