Difference between revisions of "Part:BBa K2656100"
Carolinarp (Talk | contribs) |
Carolinarp (Talk | contribs) |
||
| (2 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | |||
__NOTOC__ | __NOTOC__ | ||
<partinfo>BBa_K2656100 short</partinfo> | <partinfo>BBa_K2656100 short</partinfo> | ||
Constitutive expressed transcriptional unit assembled with a one-pot [http://2018.igem.org/Team:Valencia_UPV/Design#Level1 Level 1] Golden Gate reaction using BsaI type IIS endonuclease. | Constitutive expressed transcriptional unit assembled with a one-pot [http://2018.igem.org/Team:Valencia_UPV/Design#Level1 Level 1] Golden Gate reaction using BsaI type IIS endonuclease. | ||
| + | |||
| + | This composite part of our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection#com '''BioArt DNA toolkit palette: Printone '''], as it can be used to express a green bright color with low intensity. | ||
| + | |||
This transcriptional unit is composed of the following standardized parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]: | This transcriptional unit is composed of the following standardized parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]: | ||
| Line 15: | Line 17: | ||
</html> | </html> | ||
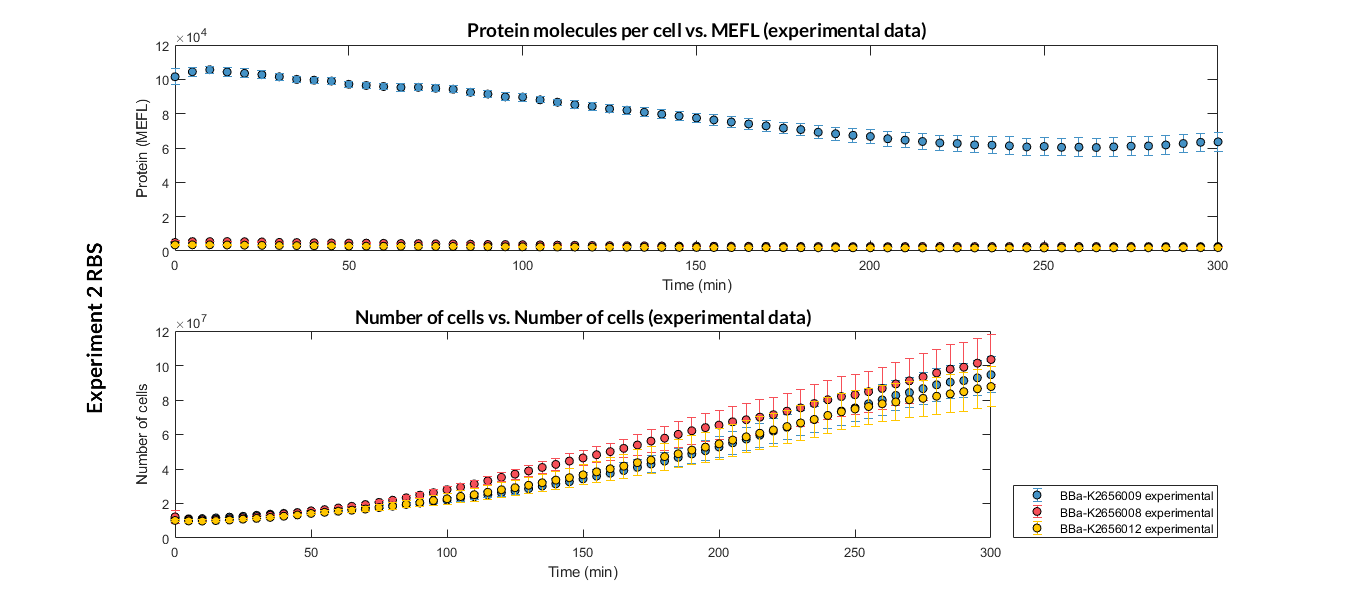
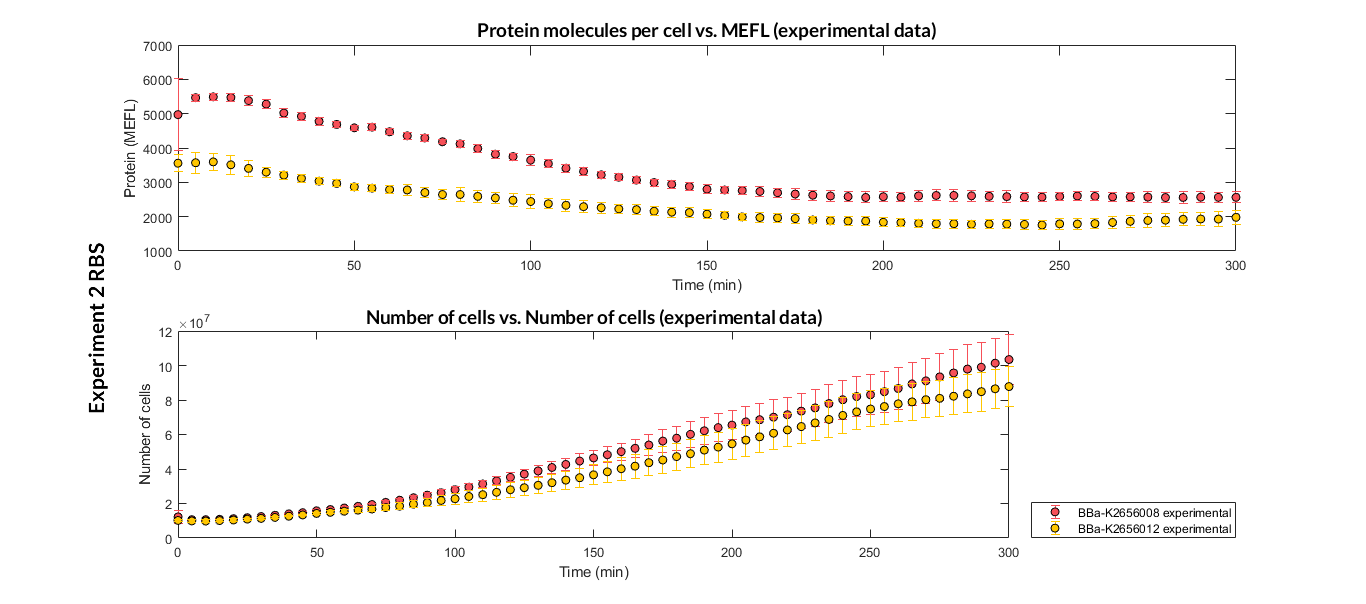
| − | This transcriptional unit has been used to characterize the RBS basic part [https://parts.igem.org/Part:BBa_K2656008 BBa_K2656008]. Following this [http://2018.igem.org/Team:Valencia_UPV/Modeling#http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model] and the results of the next graphs have been obtained after an optimization and decision-making process, in which the optimal parameters have been selected and the constituent model for each objective has been simulated and compared with the experimental data. | + | This transcriptional unit has been used to characterize the RBS basic part [https://parts.igem.org/Part:BBa_K2656008 BBa_K2656008]. Following this [http://2018.igem.org/Team:Valencia_UPV/Modeling#http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model] and the results of the next graphs have been obtained after an [http://2018.igem.org/Team:Valencia_UPV/Modeling#optimization optimization] and decision-making process, in which the optimal parameters have been selected and the constituent model for each objective has been simulated and compared with the experimental data. |
| − | [[File:T--Valencia_UPV--optimization_exp2_RBS_all_graph_expUPV2018.png|900px|thumb|none|alt=RBS experiment.|Figure 1. RBS expression experiment | + | [[File:T--Valencia_UPV--optimization_exp2_RBS_all_graph_expUPV2018.png|900px|thumb|none|alt=RBS experiment.|Figure 1. RBS expression experiment of K2656008, K2656009 and K2656012 basic pieces]] |
| − | [[File:T--Valencia_UPV--optimization_exp2_RBS_2obj_graph_expUPV2018.png|900px|thumb|none|alt=RBS experiment.|Figure 2. RBS expression experiment | + | [[File:T--Valencia_UPV--optimization_exp2_RBS_2obj_graph_expUPV2018.png|900px|thumb|none|alt=RBS experiment.|Figure 2. RBS expression experiment of K2656008 and K2656012 basic pieces]] |
Latest revision as of 08:30, 16 October 2018
sfGFP transcriptional unit 1
Constitutive expressed transcriptional unit assembled with a one-pot [http://2018.igem.org/Team:Valencia_UPV/Design#Level1 Level 1] Golden Gate reaction using BsaI type IIS endonuclease.
This composite part of our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection#com BioArt DNA toolkit palette: Printone ], as it can be used to express a green bright color with low intensity.
This transcriptional unit is composed of the following standardized parts from our [http://2018.igem.org/Team:Valencia_UPV/Part_Collection Part Collection]:
- BBa_K2656004: the J23106 constitutive promoter in its Golden Braid compatible version
- BBa_K2656008: the J61100 very weak ribosome biding site in its Golden Braid compatible version
- BBa_K2656013: the BBa_I746916 sfGFP sequence in its Golden Braid standardized version
- Terminator BBa_K2656026: the B0015 transcriptional terminator in its Golden Braid compatible version
This transcriptional unit has been used to characterize the RBS basic part BBa_K2656008. Following this [http://2018.igem.org/Team:Valencia_UPV/Modeling#http://2018.igem.org/Team:Valencia_UPV/Experiments#exp_protocol experimental protocol], we have obtained the parameters to valide our [http://2018.igem.org/Team:Valencia_UPV/Modeling#models constitutive model] and the results of the next graphs have been obtained after an [http://2018.igem.org/Team:Valencia_UPV/Modeling#optimization optimization] and decision-making process, in which the optimal parameters have been selected and the constituent model for each objective has been simulated and compared with the experimental data.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 11
Illegal NheI site found at 34 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 77