Difference between revisions of "Part:BBa K2571006"
| Line 30: | Line 30: | ||
| − | <table> | + | <table style = "border = 1px solid black"> |
<tr> | <tr> | ||
<td> Kutay </td> | <td> Kutay </td> | ||
Revision as of 05:45, 16 October 2018
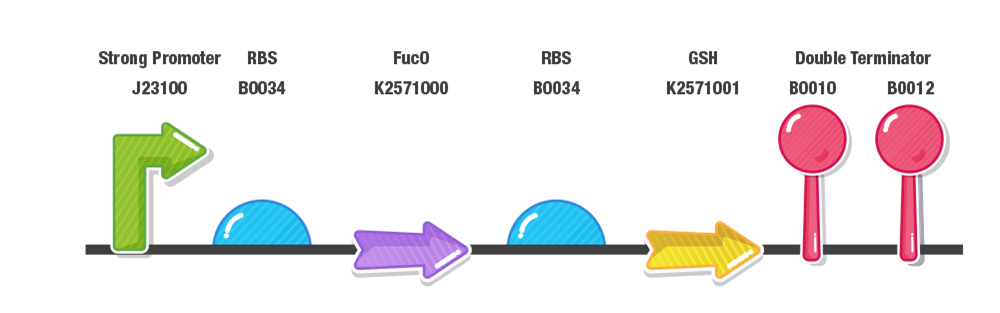
Dual Expression of FucO and GSH
Usage and Biology
The first protein coding region we have, placed after the RBS, FucO, will code for L-1,2-propanediol oxidoreductase (a homodimer enzyme) in order to act upon furfural presence in the field (Zheng, 2013). The metabolism of furfural by NAD(P)H-dependent oxidoreductases will reduce the toxicity of the chemical by turning it into furfuryl alcohol, a derivative and increase the furfural tolerance (Zheng, 2013; Wang et al., 2013; Allen et al., 2010). Our second protein coding region, bifunctional gamma-glutamate-cysteine ligase/glutathione synthetase (GSH), is a non-protein thiol group and a tripeptide composed of cysteine, glycine and glutamic acid (Lu, 2013). It is crucial for the detoxification of reactive oxygen species and free radicals (Ask et al, 2013). Reactive oxygen species (ROS) are harmful substances that alter protein based matters by taking electrons (Lu, 2013; Burton & Jauniaux, 2011). Because many benefits of GSH include scavenging of ROS, protection against endogenous toxic metabolites and detoxification of xenobiotics, we choose this gene to entagrate with the FucO (Höck et al., 2013). Thus we constructed multi functional gene providing long life span and resistance.
Design Notes of Dual Expression of FucO and GSH (BBa_K2571006)
Our construct for composite part 3 is composed of two stages, first the reduction of furans (specifically furfural and 5-HMF) and second the detoxification of reactive oxygen species (ROS). To achieve this effect, we designed our composite 3 part as with a prefix, a strong promoter (J23100), RBS (B0034), fucO as the first protein coding region (BBa_K2571003), RBS (B0034), GSH as the second protein coding region(BBa_K2571005) , double terminator (B0015) and suffix.
Our construct is inserted into pSB1C3 and delivered to the Registry. Our construct is also inserted into pSB1A3 and transferred into KO11 to conduct further biochemical assays.
Given that fucO is NADH-dependent it outperforms other oxidoreductases, by not interfering with the NADPH metabolism of the organism while converting highly toxic substances, furfural and 5-HMF to non-harmful alcohols. This characteristic of fucO is crucial because the expression of oxidoreductases like Yqhd are NADPH-dependent, hence they compete with the biosynthesis for NADPH, which results in inhibiting the growth of the organism.
Glutathione, on the other hand, is recycled using NAD(P)H pathways and since now it will be overexpressed and with NADH metabolism is not being altered thanks to FucO, antioxidant capacity of the cell will be increased dramatically, result in amplified immunity to both furans and ROS, habilitating cell growth, increasing ethanol yield by the virtue of increasing cell mass and reproduction, and improved metabolism.
| Kutay | Garip |
Gel Characterization
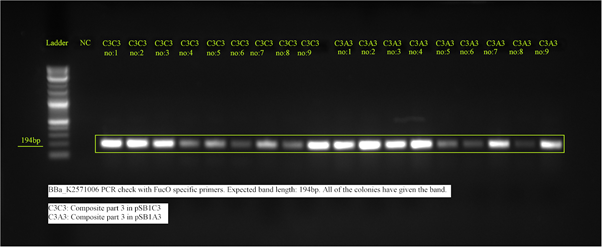
We’ve inserted our composite part 3(BBa_K2571006) in both pSB1C3 and pSB1A3 backbones. The construct in pSB1C3 is for submission to registry and is cultivated in DH5 alpha. The plasmid having pSB1A3 as backbone, thus carrying ampicillin resistance is for our biochemical assays since we’ve chosen the chassis organism for assays as E.coli strain KO11 which already has Chloramphenicol resistance in its genome. After cloning our genes, we’ve made colony PCR to verify our insertions. We chose the primers as FucO specific since the composite 3 contains FucO coding region. Expected band length was 194 bp, and as expected, the bands were given by all of the DH5 alpha and KO11 colonies we chose, confirming our transformations.
FucO specific primers were used:
FucO left: GTGATAAGGATGCCGGAGAA
FucO right: CTTCTCGCCGGTAAAGTCAG
References
Allen, S. A., Clark, W., McCaffery, J. M., Cai, Z., Lanctot, A., Slininger, P. J., … Gorsich, S. W. (2010). Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnology for Biofuels, 3, 2. http://doi.org/10.1186/1754-6834-3-2
Ask, M., Mapelli, V., Höck, H., Olsson, L., Bettiga, M. (2013) Engineering glutathione biosynthesis of Saccharomyces cerevisiae increases robustness to inhibitors in pretreated lignocellulosic materials. Microbial Cell Factories. 12:87 https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3817835/
Burton, G. J., & Jauniaux, E. (2011). Oxidative stress. Best Practice & Research. Clinical Obstetrics & Gynaecology, 25(3), 287–299. http://doi.org/10.1016/j.bpobgyn.2010.10.016
Chou, H.-H., Marx, C. J., & Sauer, U. (2015). Transhydrogenase Promotes the Robustness and Evolvability of E. coli Deficient in NADPH Production. PLoS Genetics, 11(2), e1005007. http://doi.org/10.1371/journal.pgen.1005007
Liu, Z.L., Ma M., Song, M.(2009). Evolutionarily engineered ethanologenic yeast detoxifies lignocellulosic biomass conversion inhibitors by reprogrammed pathways. Mol Genet Genomics 282, 233-244. doi: 10.1007/s00438-009-0461-7
Lu, S. C. (2013). GLUTATHIONE SYNTHESIS. Biochemica et Biophysica Acta, 1830(5), 3143–3153. http://doi.org/10.1016/j.bbagen.2012.09.008
National Center for Biotechnology Information. PubChem Compound Database; CID=124886, https://pubchem.ncbi.nlm.nih.gov/compound/124886 (accessed July 18, 2018). https://pubchem.ncbi.nlm.nih.gov/compound/124886#section=Top
Patrick, L. (2003). Mercury Toxicity and Antioxidants: Part I: Role of Glutathione and alpha-Lipoic Acid in the Treatment of Mercury Toxicity. Alternative medicine review: a journal of clinical therapeutic.(7). 456-471. https://www.researchgate.net/publication/10980025_Mercury_Toxicity_and_Antioxidants_Part_I_Role_of_Glutathione_and_alpha-Lipoic_Acid_in_the_Treatment_of_Mercury_Toxicity
Pizzorno, J. (2014). Glutathione! Integrative Medicine: A Clinician’s Journal, 13(1), 8–12. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4684116/
Wang, X., Miller, E. N., Yomano, L. P., Zhang, X., Shanmugam, K. T., & Ingram, L. O. (2011). Increased Furfural Tolerance Due to Overexpression of NADH-Dependent Oxidoreductase FucO in Escherichia coli Strains Engineered for the Production of Ethanol and Lactate. Applied and Environmental Microbiology, 77(15), 5132–5140. http://doi.org/10.1128/AEM.05008-11
Wang, X., Yomano, L. P., Lee, J. Y., York, S. W., Zheng, H., Mullinnix, M. T., … Ingram, L. O. (2013). Engineering furfural tolerance in Escherichia coli improves the fermentation of lignocellulosic sugars into renewable chemicals. Proceedings of the National Academy of Sciences of the United States of America, 110(10), 4021–4026. http://doi.org/10.1073/pnas.1217958110
Zheng, H., Wang, X., Yomano, L.P., Geddes, R. D, Shanmugan, K. T., Ingram, L.O. (2013). Improving Escherichia coli FucO for Furfural Tolerance by Saturation Mutagenesis of Individual Amino Acid Positions. Applied and Environmental Microbiology Vol 79, no 10. 3202–3208. http://aem.asm.org/content/79/10/3202.full.pdf+html
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12INCOMPATIBLE WITH RFC[12]Illegal NheI site found at 7
Illegal NheI site found at 30 - 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal BsaI.rc site found at 3247