Difference between revisions of "Part:BBa K2549007"
(→Usage and Biology) |
m |
||
| (3 intermediate revisions by the same user not shown) | |||
| Line 4: | Line 4: | ||
This part is the mouse Notch1 extended regulatory region<ref>Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Morsut L, Roybal KT, Xiong X, ..., Thomson M, Lim WA. Cell, 2016 Feb;164(4):780-91 PMID: 26830878; DOI: 10.1016/j.cell.2016.01.012</ref>. It was used as the transmembrane core domain of the SynNotch. However, original reported design has a high background activation in transfected 293T cells. We have put a lot of effect in optimizing this critical transmembrane domain for Notch receptor. Comparing with [[Part:BBa_K2549006]], minimal regulatory region (Ile1427 to Arg1752), extended regulatory region (Pro1390 to Arg1752) includes additional '''3''' EGF repeats. | This part is the mouse Notch1 extended regulatory region<ref>Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Morsut L, Roybal KT, Xiong X, ..., Thomson M, Lim WA. Cell, 2016 Feb;164(4):780-91 PMID: 26830878; DOI: 10.1016/j.cell.2016.01.012</ref>. It was used as the transmembrane core domain of the SynNotch. However, original reported design has a high background activation in transfected 293T cells. We have put a lot of effect in optimizing this critical transmembrane domain for Notch receptor. Comparing with [[Part:BBa_K2549006]], minimal regulatory region (Ile1427 to Arg1752), extended regulatory region (Pro1390 to Arg1752) includes additional '''3''' EGF repeats. | ||
| − | |||
<span class='h3bb'>Sequence and Features</span> | <span class='h3bb'>Sequence and Features</span> | ||
| Line 11: | Line 10: | ||
<!-- Add more about the biology of this part here --> | <!-- Add more about the biology of this part here --> | ||
| + | ===Biology=== | ||
| + | =====Significance of Notch signaling ===== | ||
| + | The Notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms. Mammals possess four different notch receptors, referred to as NOTCH1 to NOTCH4. The notch receptor is a single-pass transmembrane receptor protein<ref>https://en.wikipedia.org/wiki/Notch_signaling_pathway</ref>. | ||
| + | |||
| + | [[File:Notch-signal.png|none|400px|thumb|''Notch signaling is an evolutionarily conserved pathway in multicellular organisms that regulates cell-fate determination during development and maintains adult tissue homeostasis. The Notch pathway mediates juxtacrine cellular signaling wherein both the signal sending and receiving cells are affected through ligand-receptor crosstalk by which an array of cell fate decisions in neuronal, cardiac, immune, and endocrine development are regulated''<ref>https://www.cellsignal.com/contents/science-cst-pathways-stem-cell-markers/notch-signaling-interactive-pathway/pathways-notch</ref>.]] | ||
| + | |||
| + | Dr. Hans Widlund (Brigham and Women’s Hospital, Harvard Medical School) once contributed a list (shown below) of original articles worth reading. There are significant amount of Notch research articles and reviews in PubMed<ref>https://www.ncbi.nlm.nih.gov/pubmed/?term=notch</ref> as well. | ||
| + | * Ables JL, Breunig JJ, Eisch AJ, Rakic P (2011) Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12(5), 269–83. | ||
| + | * Andersson ER, Lendahl U (2014) Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov 13(5), 357–78. | ||
| + | * Aster JC, Blacklow SC, Pear WS (2011) Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223(2), 262–73. | ||
| + | * Bai G, Pfaff SL (2011) Protease regulation: the Yin and Yang of neural development and disease. Neuron 72(1), 9–21. | ||
| + | * de la Pompa JL, Epstein JA (2012) Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22(2), 244–54. | ||
| + | * Ntziachristos P, Lim JS, Sage J, Aifantis I (2014) From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25(3), 318–34. | ||
| + | * Ranganathan P, Weaver KL, Capobianco AJ (2011) Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11(5), 338–51. | ||
| + | * Weinmaster G, Fischer JA (2011) Notch ligand ubiquitylation: what is it good for? Dev. Cell 21(1), 134–44. | ||
| + | * Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ (2010) Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 28, 343–65. | ||
| + | |||
===== synNotch with α-CD19 against CD19 antigen works extremely well in Morsut L et al 2016===== | ===== synNotch with α-CD19 against CD19 antigen works extremely well in Morsut L et al 2016===== | ||
Please refer the original article for more details. | Please refer the original article for more details. | ||
| Line 35: | Line 51: | ||
Note: | Note: | ||
| − | + | [[Part:BBa_K2549005]] is used in the study described above (Morsut L et al). | |
[[Part:BBa_K2549007]] and [[Part:BBa_K2549008]] have the same Biology section. Our experiments suggested mouse Notch 1 core '''without extended 3 EGF repeats''' works better and our receptor optimization all uses [[Part:BBa_K2549006]]. | [[Part:BBa_K2549007]] and [[Part:BBa_K2549008]] have the same Biology section. Our experiments suggested mouse Notch 1 core '''without extended 3 EGF repeats''' works better and our receptor optimization all uses [[Part:BBa_K2549006]]. | ||
<!-- Uncomment this to enable Functional Parameter display | <!-- Uncomment this to enable Functional Parameter display | ||
===Functional Parameters=== | ===Functional Parameters=== | ||
| − | <partinfo> | + | <partinfo>BBa_K2549007 parameters</partinfo> |
--> | --> | ||
===References=== | ===References=== | ||
Latest revision as of 14:34, 7 October 2018
mouse Notch1 core (extended)
This part is the mouse Notch1 extended regulatory region[1]. It was used as the transmembrane core domain of the SynNotch. However, original reported design has a high background activation in transfected 293T cells. We have put a lot of effect in optimizing this critical transmembrane domain for Notch receptor. Comparing with Part:BBa_K2549006, minimal regulatory region (Ile1427 to Arg1752), extended regulatory region (Pro1390 to Arg1752) includes additional 3 EGF repeats.
Sequence and Features
- 10COMPATIBLE WITH RFC[10]
- 12COMPATIBLE WITH RFC[12]
- 21COMPATIBLE WITH RFC[21]
- 23COMPATIBLE WITH RFC[23]
- 25COMPATIBLE WITH RFC[25]
- 1000INCOMPATIBLE WITH RFC[1000]Illegal SapI.rc site found at 703
Biology
Significance of Notch signaling
The Notch signaling pathway is a highly conserved cell signaling system present in most multicellular organisms. Mammals possess four different notch receptors, referred to as NOTCH1 to NOTCH4. The notch receptor is a single-pass transmembrane receptor protein[2].
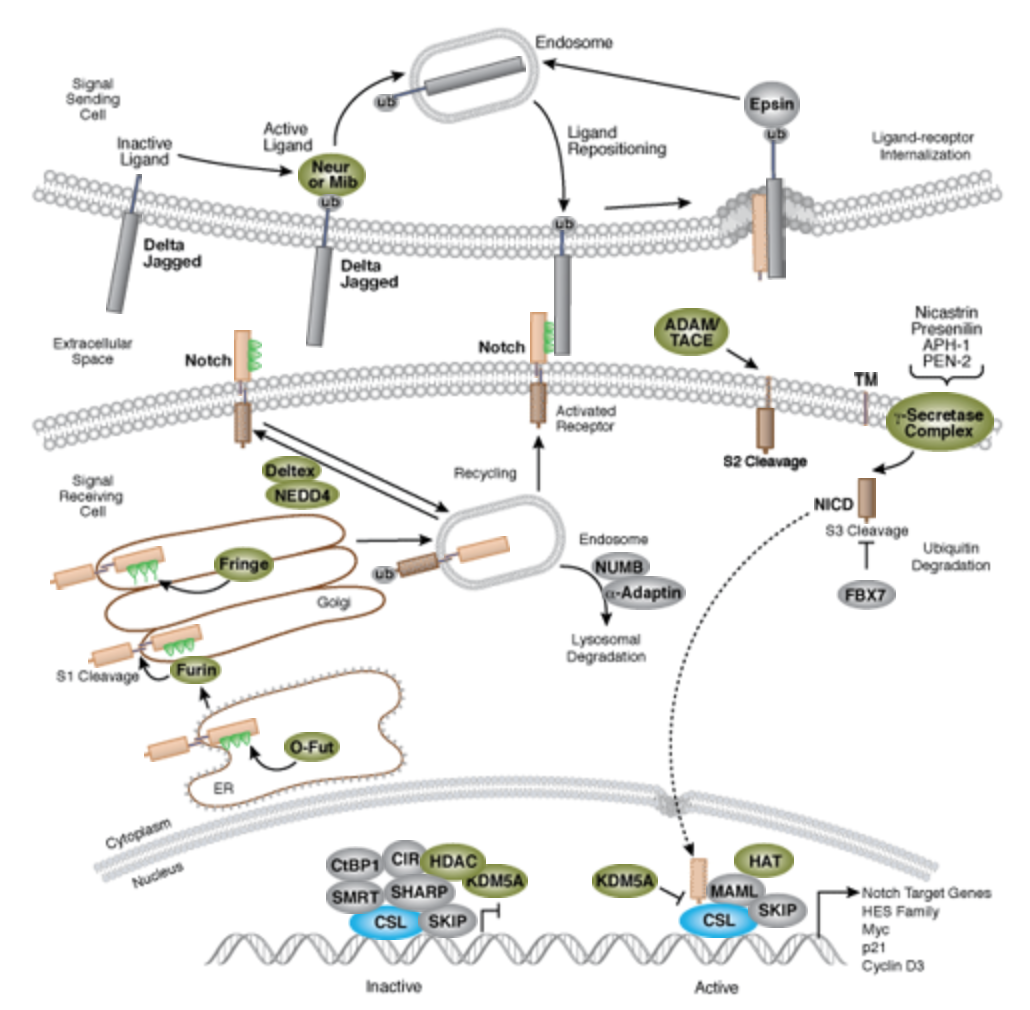
Dr. Hans Widlund (Brigham and Women’s Hospital, Harvard Medical School) once contributed a list (shown below) of original articles worth reading. There are significant amount of Notch research articles and reviews in PubMed[4] as well.
- Ables JL, Breunig JJ, Eisch AJ, Rakic P (2011) Not(ch) just development: Notch signalling in the adult brain. Nat. Rev. Neurosci. 12(5), 269–83.
- Andersson ER, Lendahl U (2014) Therapeutic modulation of Notch signalling--are we there yet? Nat Rev Drug Discov 13(5), 357–78.
- Aster JC, Blacklow SC, Pear WS (2011) Notch signalling in T-cell lymphoblastic leukaemia/lymphoma and other haematological malignancies. J. Pathol. 223(2), 262–73.
- Bai G, Pfaff SL (2011) Protease regulation: the Yin and Yang of neural development and disease. Neuron 72(1), 9–21.
- de la Pompa JL, Epstein JA (2012) Coordinating tissue interactions: Notch signaling in cardiac development and disease. Dev. Cell 22(2), 244–54.
- Ntziachristos P, Lim JS, Sage J, Aifantis I (2014) From fly wings to targeted cancer therapies: a centennial for notch signaling. Cancer Cell 25(3), 318–34.
- Ranganathan P, Weaver KL, Capobianco AJ (2011) Notch signalling in solid tumours: a little bit of everything but not all the time. Nat. Rev. Cancer 11(5), 338–51.
- Weinmaster G, Fischer JA (2011) Notch ligand ubiquitylation: what is it good for? Dev. Cell 21(1), 134–44.
- Yuan JS, Kousis PC, Suliman S, Visan I, Guidos CJ (2010) Functions of notch signaling in the immune system: consensus and controversies. Annu. Rev. Immunol. 28, 343–65.
synNotch with α-CD19 against CD19 antigen works extremely well in Morsut L et al 2016
Please refer the original article for more details.
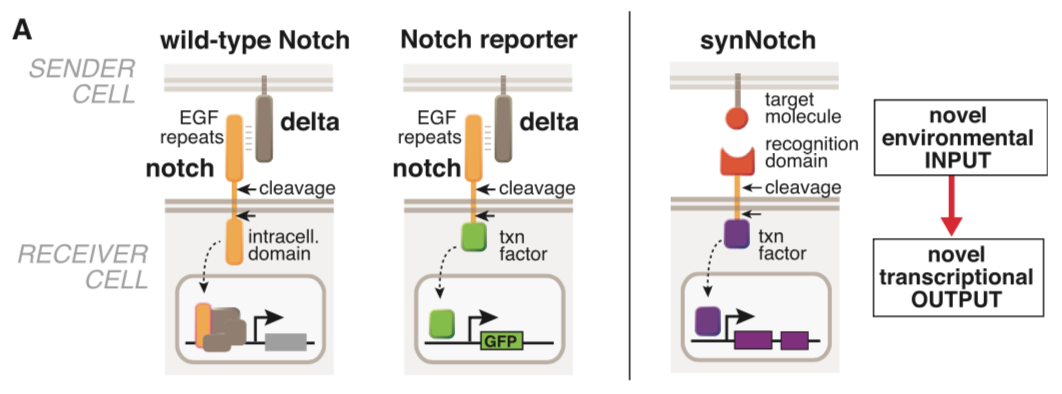
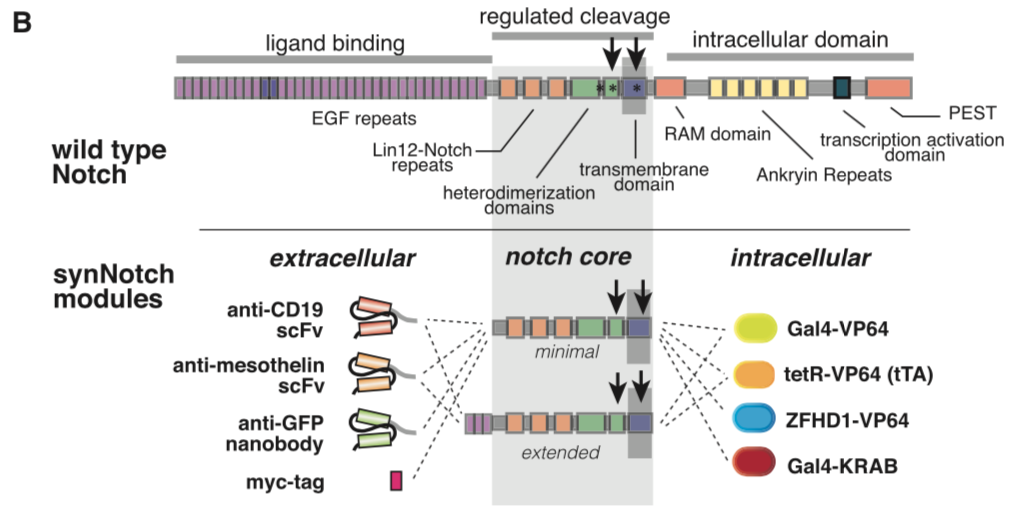





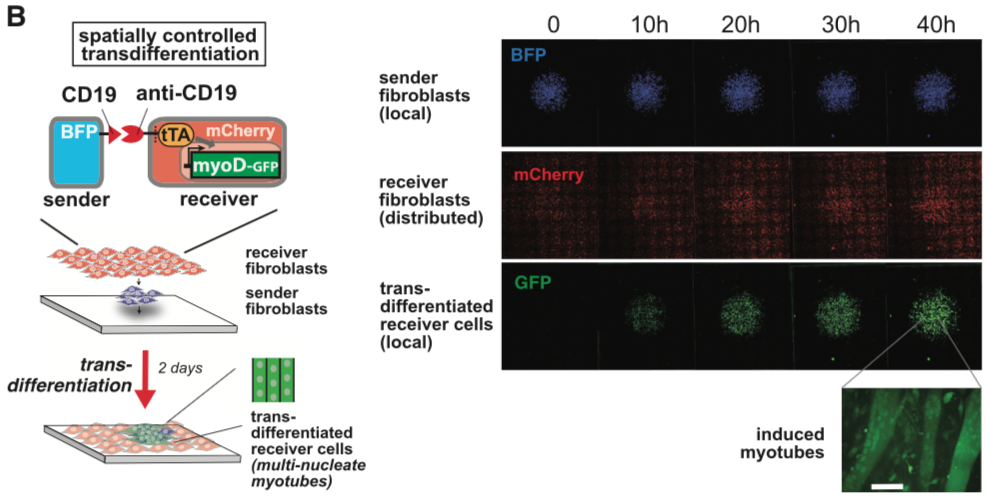

Note: Part:BBa_K2549005 is used in the study described above (Morsut L et al). Part:BBa_K2549007 and Part:BBa_K2549008 have the same Biology section. Our experiments suggested mouse Notch 1 core without extended 3 EGF repeats works better and our receptor optimization all uses Part:BBa_K2549006.
References
- ↑ Engineering Customized Cell Sensing and Response Behaviors Using Synthetic Notch Receptors. Morsut L, Roybal KT, Xiong X, ..., Thomson M, Lim WA. Cell, 2016 Feb;164(4):780-91 PMID: 26830878; DOI: 10.1016/j.cell.2016.01.012
- ↑ https://en.wikipedia.org/wiki/Notch_signaling_pathway
- ↑ https://www.cellsignal.com/contents/science-cst-pathways-stem-cell-markers/notch-signaling-interactive-pathway/pathways-notch
- ↑ https://www.ncbi.nlm.nih.gov/pubmed/?term=notch

